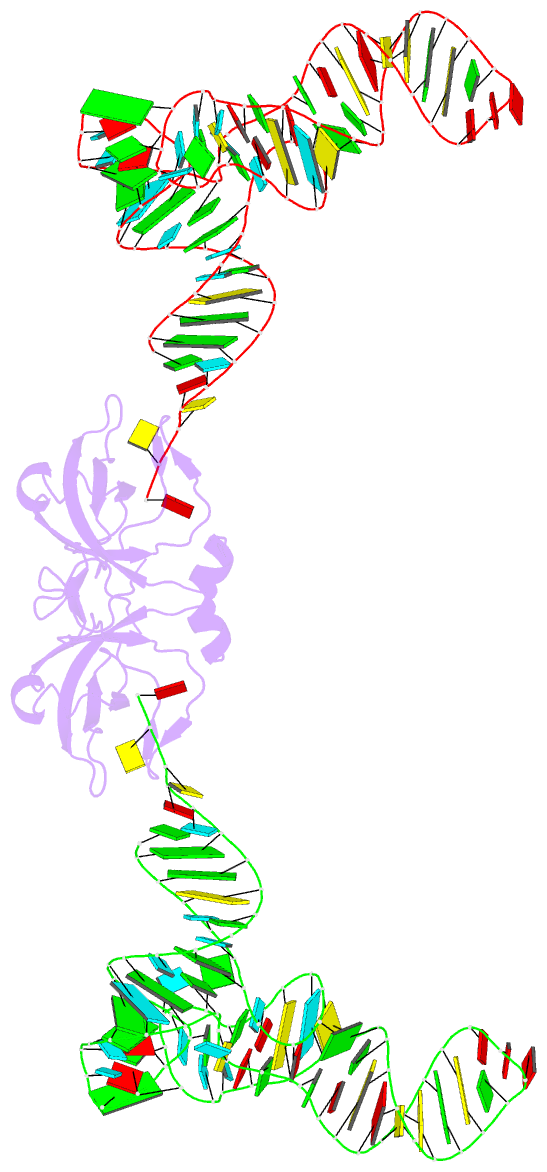
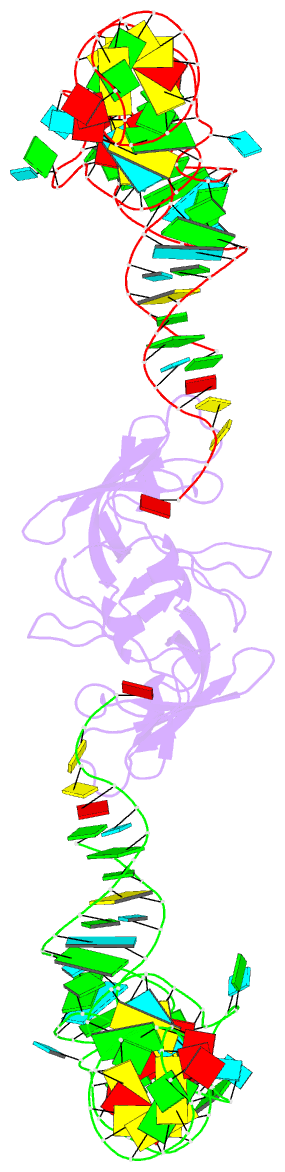
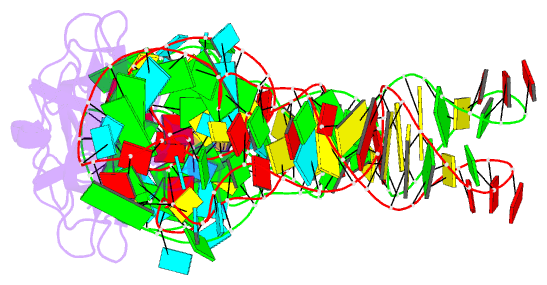
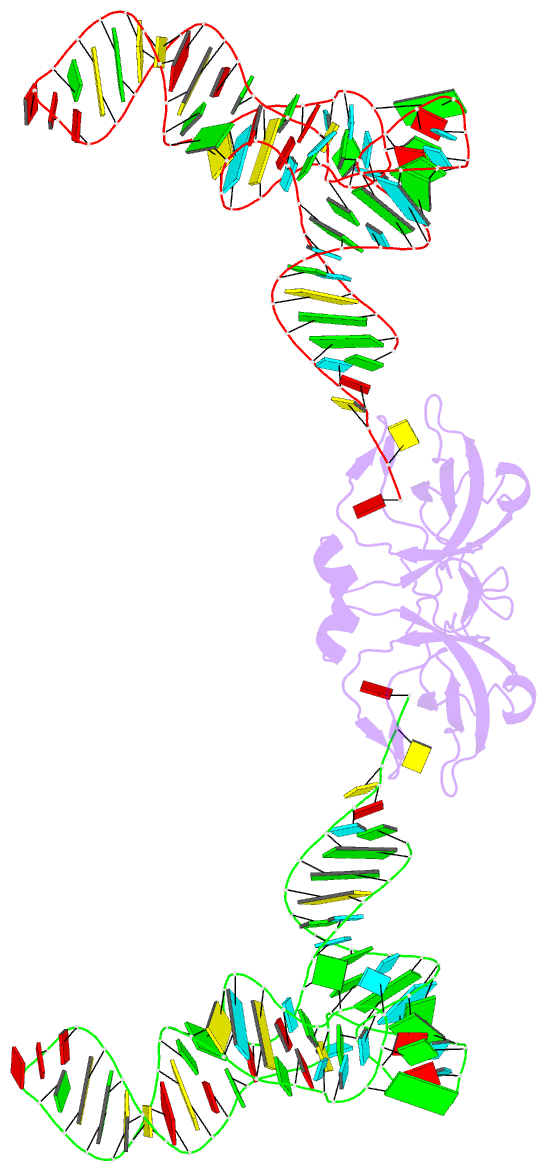
Summary information and primary citation
- PDB-id
- 8vu0; DSSR-derived features in text and JSON formats
- Class
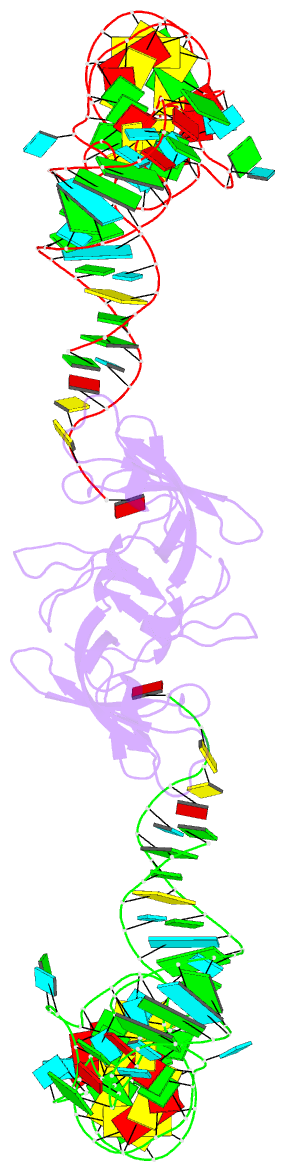
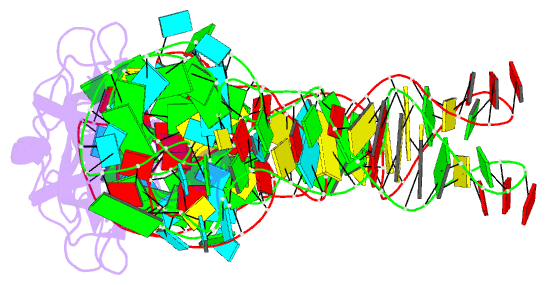
- RNA binding protein-RNA
- Method
- X-ray (2.64 Å)
- Summary
- Co-crystal structure of aquifex aeolicus trbp111 in complex with e. coli trna-ile
- Reference
- Umuhire Juru A, Ghirlando R, Zhang J (2024): "Structural basis of tRNA recognition by the widespread OB fold." Nat Commun, 15, 6385. doi: 10.1038/s41467-024-50730-1.
- Abstract
- The widespread oligonucleotide/oligosaccharide-binding (OB)-fold recognizes diverse substrates from sugars to nucleic acids and proteins, and plays key roles in genome maintenance, transcription, translation, and tRNA metabolism. OB-containing bacterial Trbp and yeast Arc1p proteins are thought to recognize the tRNA elbow or anticodon regions. Here we report a 2.6 Å co-crystal structure of Aquifex aeolicus Trbp111 bound to tRNAIle, which reveals that Trbp recognizes tRNAs solely by capturing their 3' ends. Structural, mutational, and biophysical analyses show that the Trbp/EMAPII-like OB fold precisely recognizes the single-stranded structure, 3' terminal location, and specific sequence of the 3' CA dinucleotide - a universal feature of mature tRNAs. Arc1p supplements its OB - tRNA 3' end interaction with additional contacts that involve an adjacent basic region and the tRNA body. This study uncovers a previously unrecognized mode of tRNA recognition by an ancient protein fold, and provides insights into protein-mediated tRNA aminoacylation, folding, localization, trafficking, and piracy.