Summary information and primary citation
- PDB-id
- 8tfd; DSSR-derived features in text and JSON formats
- Class
- viral protein
- Method
- X-ray (1.55 Å)
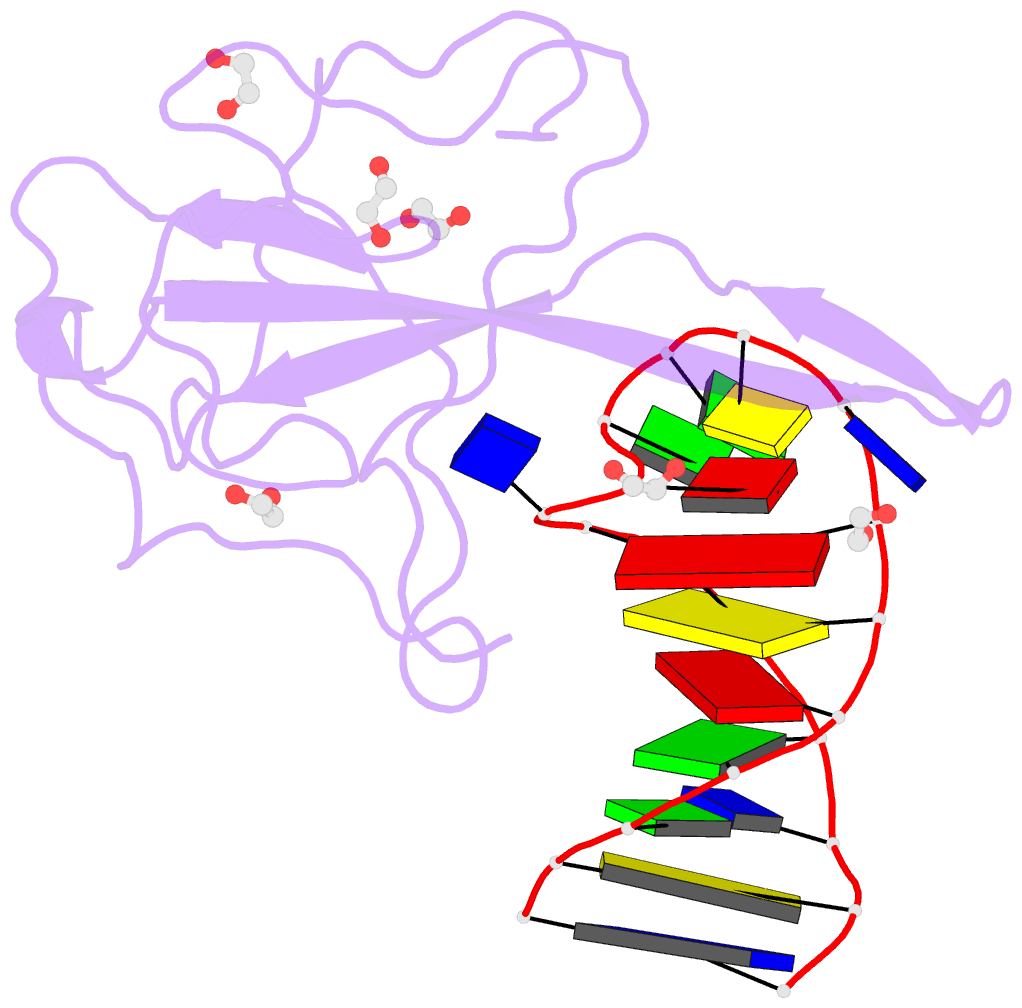
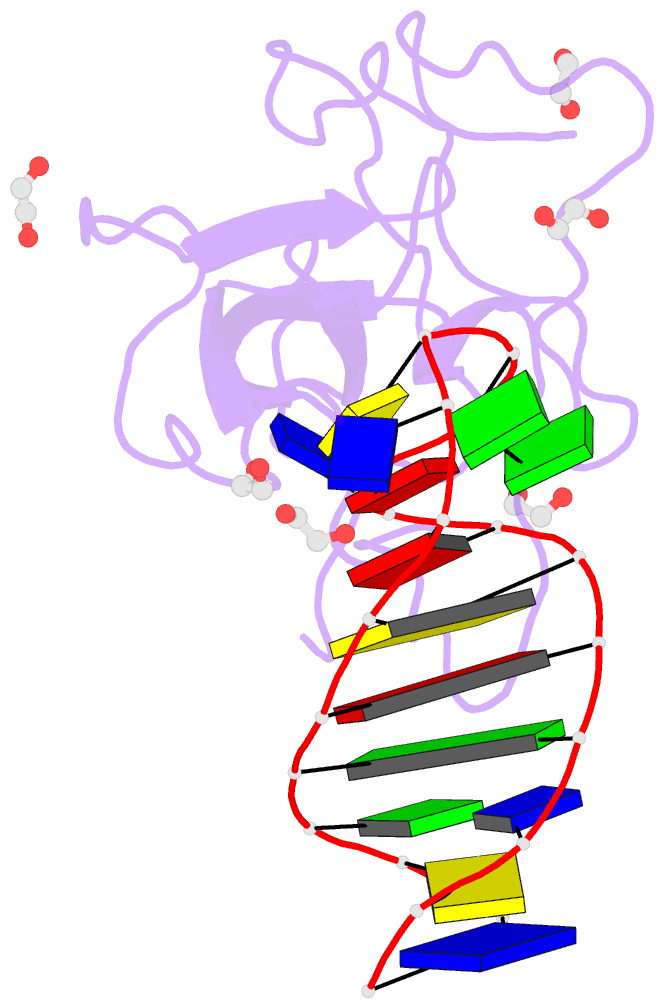
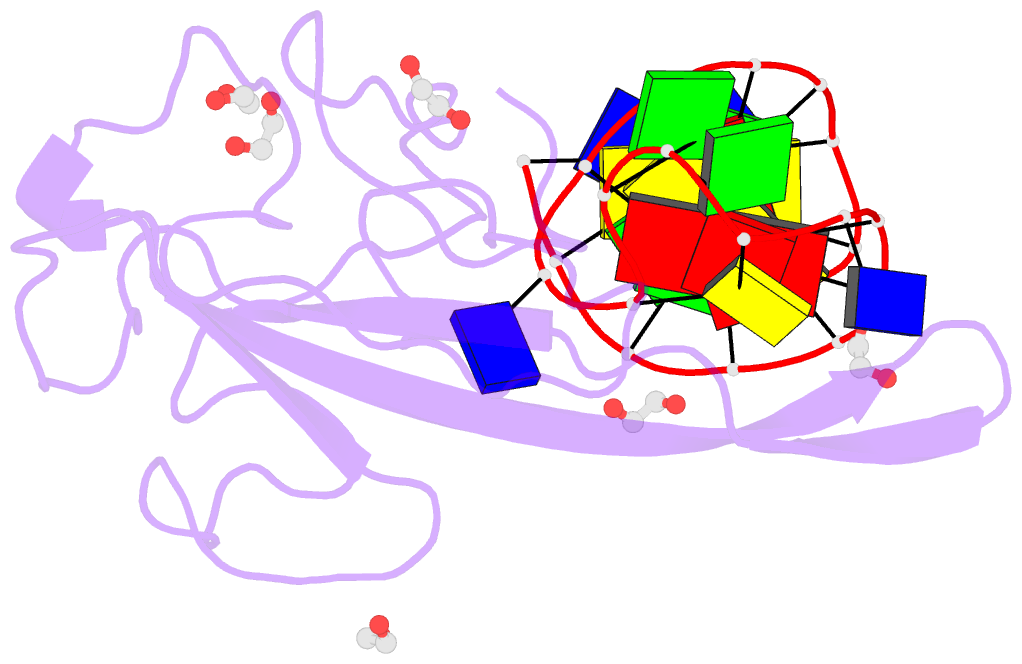
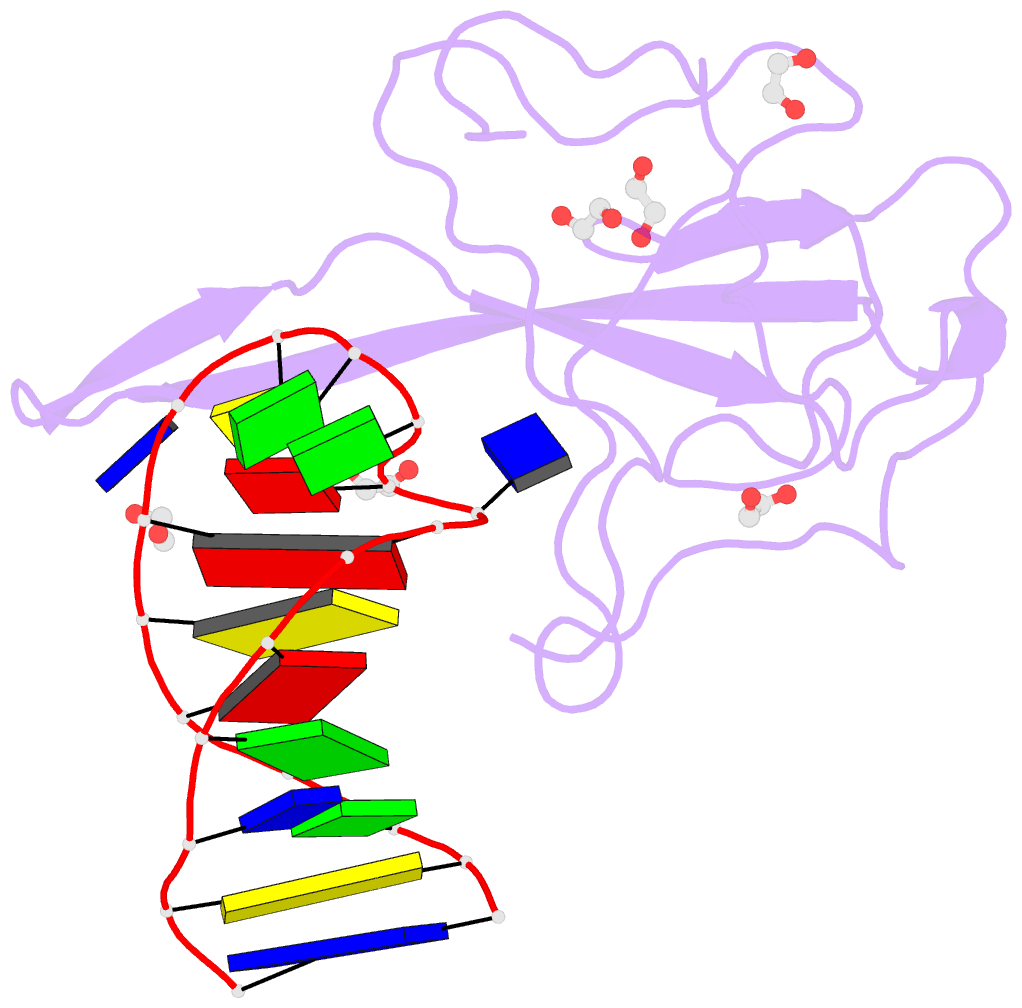
- Summary
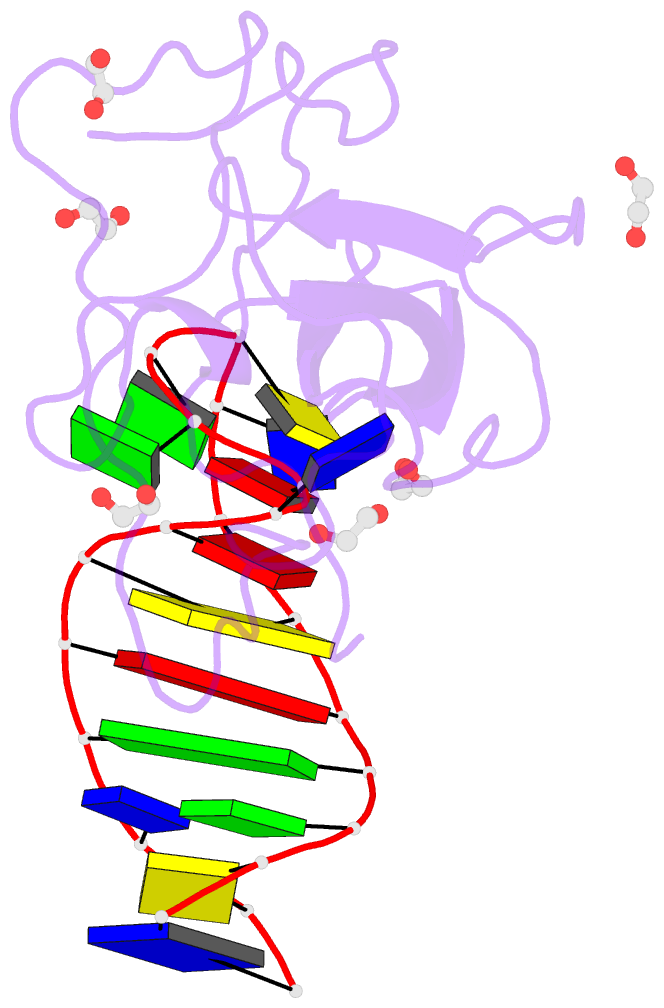
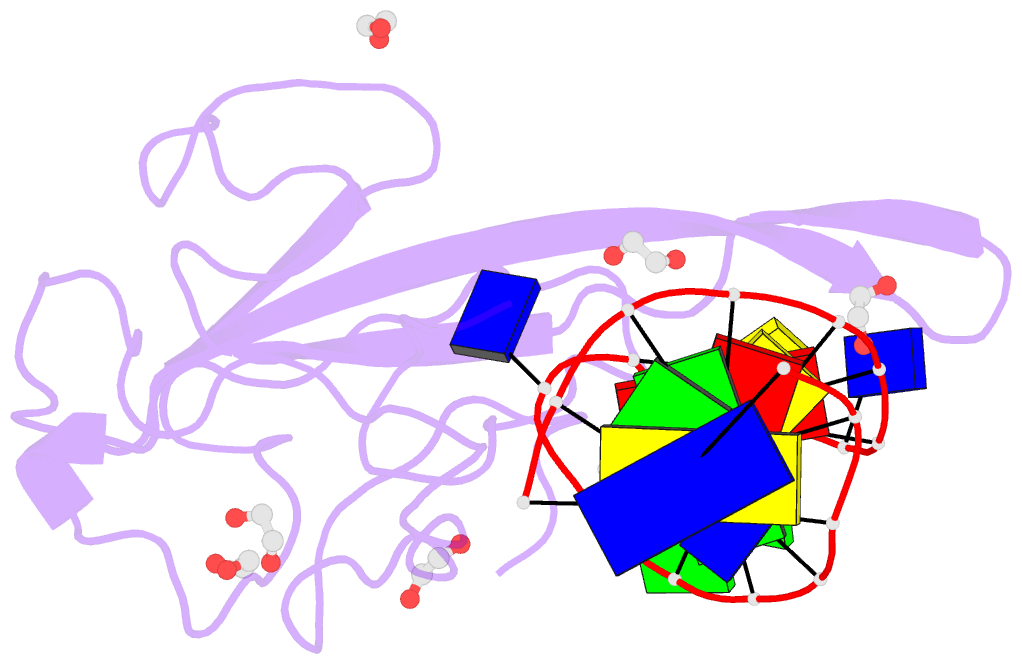
- Crystal structure of a stem-loop DNA aptamer complexed with sars-cov-2 nucleocapsid protein RNA-binding domain
- Reference
- Esler MA, Belica CA, Rollie JA, Brown WL, Moghadasi SA, Shi K, Harki DA, Harris RS, Aihara H (2024): "A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain." Nucleic Acids Res. doi: 10.1093/nar/gkae874.
- Abstract
- SARS-CoV-2 nucleocapsid (N) protein is a structural component of the virus with essential roles in the replication and packaging of the viral RNA genome. The N protein is also an important target of COVID-19 antigen tests and a promising vaccine candidate along with the spike protein. Here, we report a compact stem-loop DNA aptamer that binds tightly to the N-terminal RNA-binding domain of SARS-CoV-2 N protein. Crystallographic analysis shows that a hexanucleotide DNA motif (5'-TCGGAT-3') of the aptamer fits into a positively charged concave surface of N-NTD and engages essential RNA-binding residues including Tyr109, which mediates a sequence-specific interaction in a uracil-binding pocket. Avid binding of the DNA aptamer allows isolation and sensitive detection of full-length N protein from crude cell lysates, demonstrating its selectivity and utility in biochemical applications. We further designed a chemically modified DNA aptamer and used it as a probe to examine the interaction of N-NTD with various RNA motifs, which revealed a strong preference for uridine-rich sequences. Our studies provide a high-affinity chemical probe for the SARS-CoV-2 N protein RNA-binding domain, which may be useful for diagnostic applications and investigating novel antiviral agents.