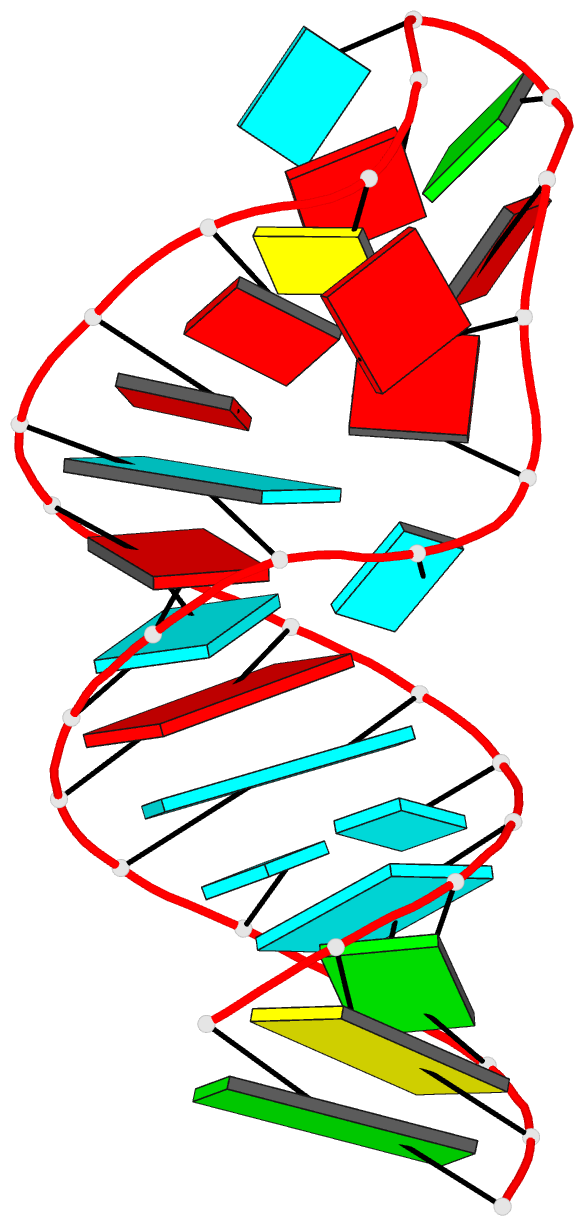
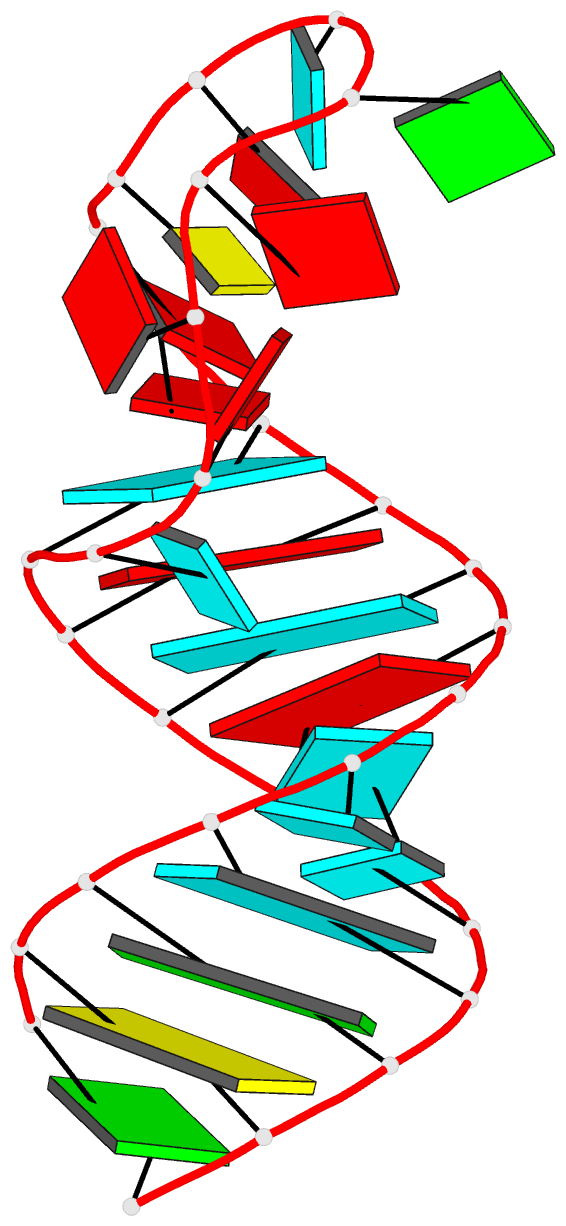
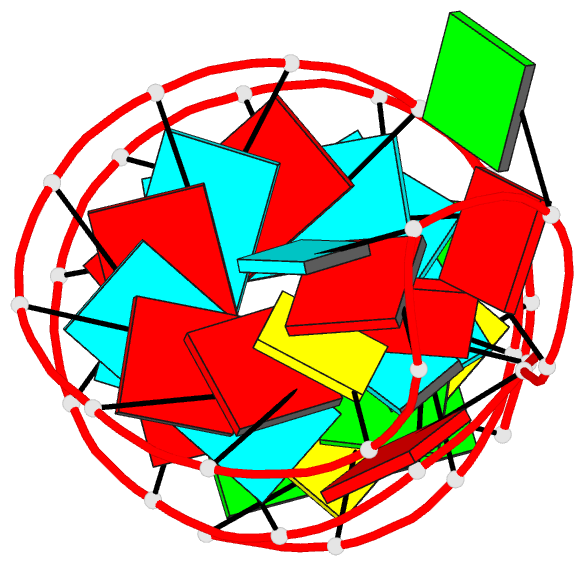
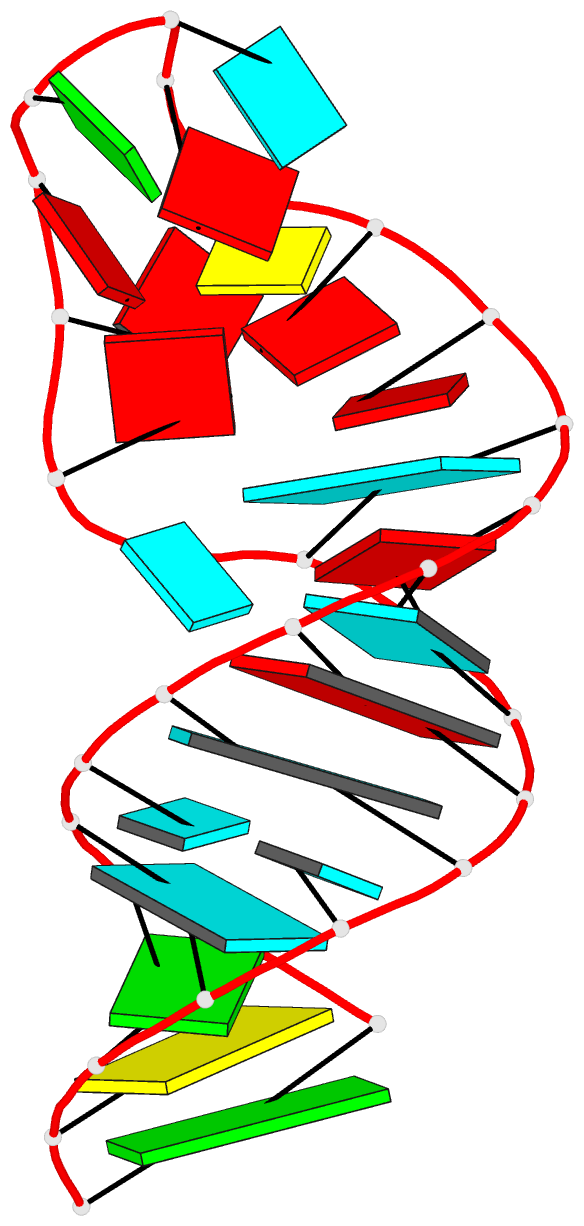
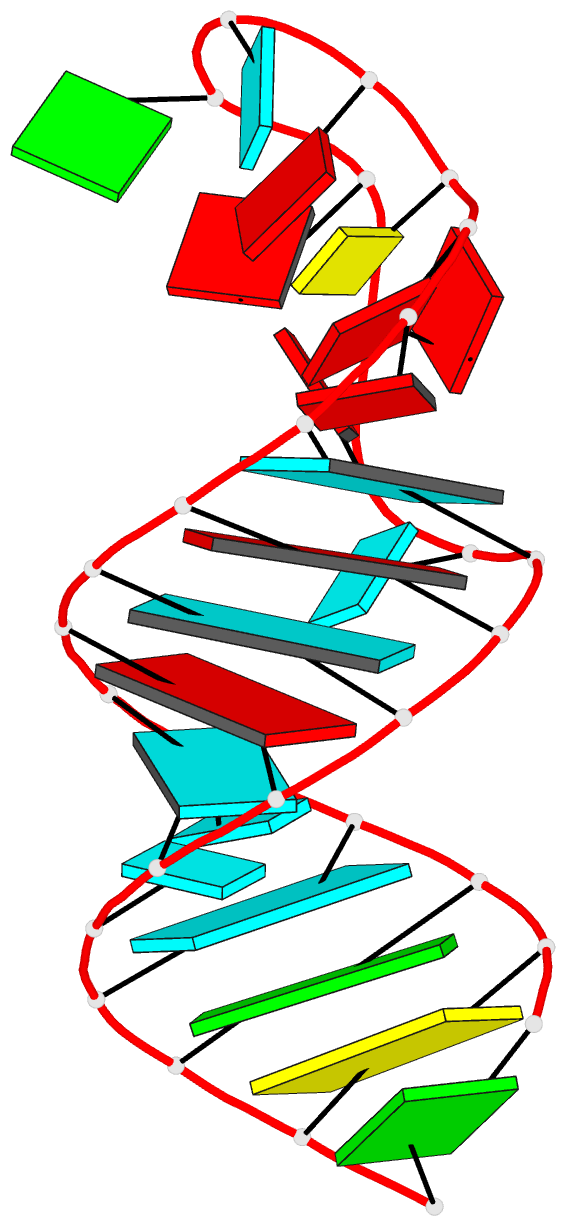
Summary information and primary citation
- PDB-id
- 8scf; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Tceiii NMR structure
- Reference
- Warden MS, DeRose EF, Tamayo JV, Mueller GA, Gavis ER, Hall TMT (2023): "The translational repressor Glorund uses interchangeable RNA recognition domains to recognize Drosophila nanos." Nucleic Acids Res. doi: 10.1093/nar/gkad586.
- Abstract
- The Drosophila melanogaster protein Glorund (Glo) represses nanos (nos) translation and uses its quasi-RNA recognition motifs (qRRMs) to recognize both G-tract and structured UA-rich motifs within the nos translational control element (TCE). We showed previously that each of the three qRRMs is multifunctional, capable of binding to G-tract and UA-rich motifs, yet if and how the qRRMs combine to recognize the nos TCE remained unclear. Here we determined solution structures of a nos TCEI_III RNA containing the G-tract and UA-rich motifs. The RNA structure demonstrated that a single qRRM is physically incapable of recognizing both RNA elements simultaneously. In vivo experiments further indicated that any two qRRMs are sufficient to repress nos translation. We probed interactions of Glo qRRMs with TCEI_III RNA using NMR paramagnetic relaxation experiments. Our in vitro and in vivo data support a model whereby tandem Glo qRRMs are indeed multifunctional and interchangeable for recognition of TCE G-tract or UA-rich motifs. This study illustrates how multiple RNA recognition modules within an RNA-binding protein may combine to diversify the RNAs that are recognized and regulated.