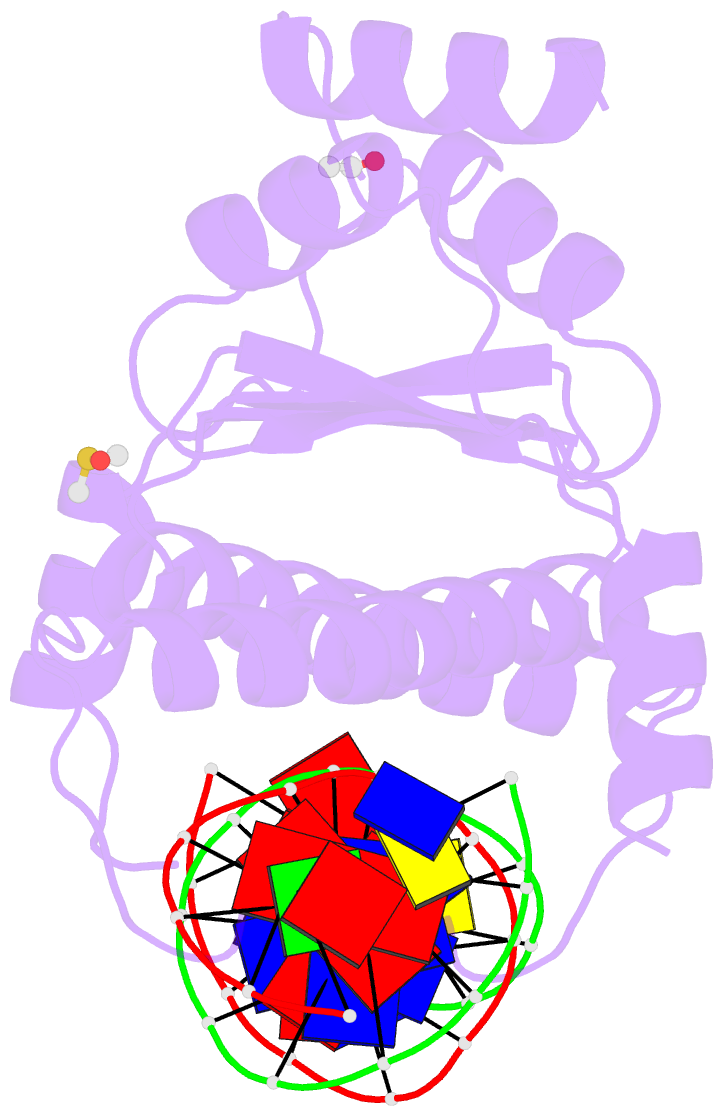
Summary information and primary citation
- PDB-id
-
8q9n;
SNAP-derived features in text and
JSON formats
- Class
- transcription
- Method
- X-ray (1.51 Å)
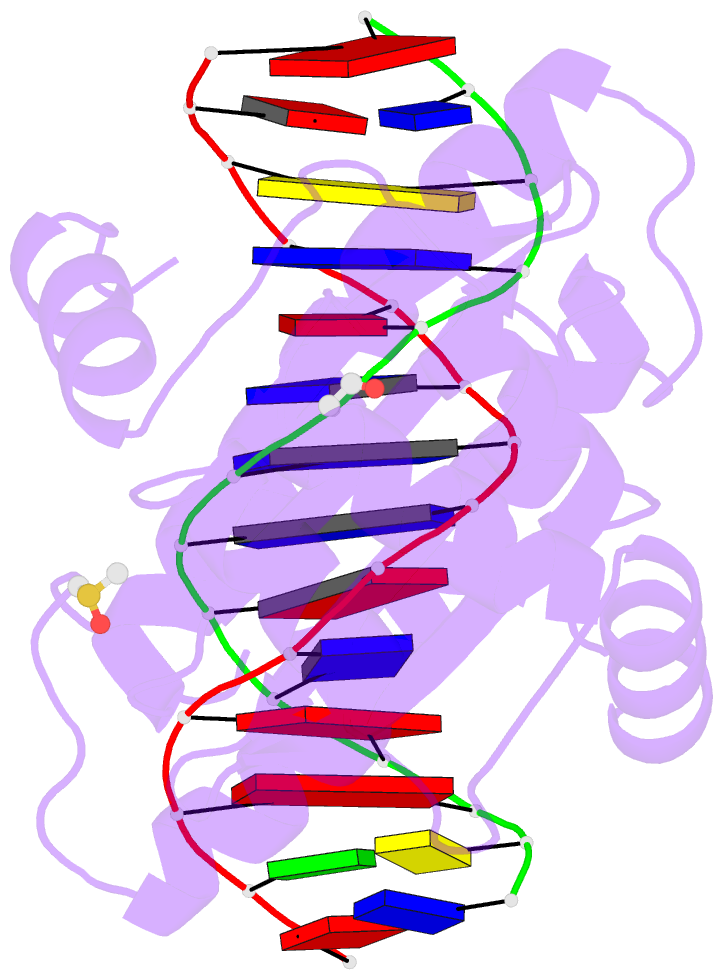
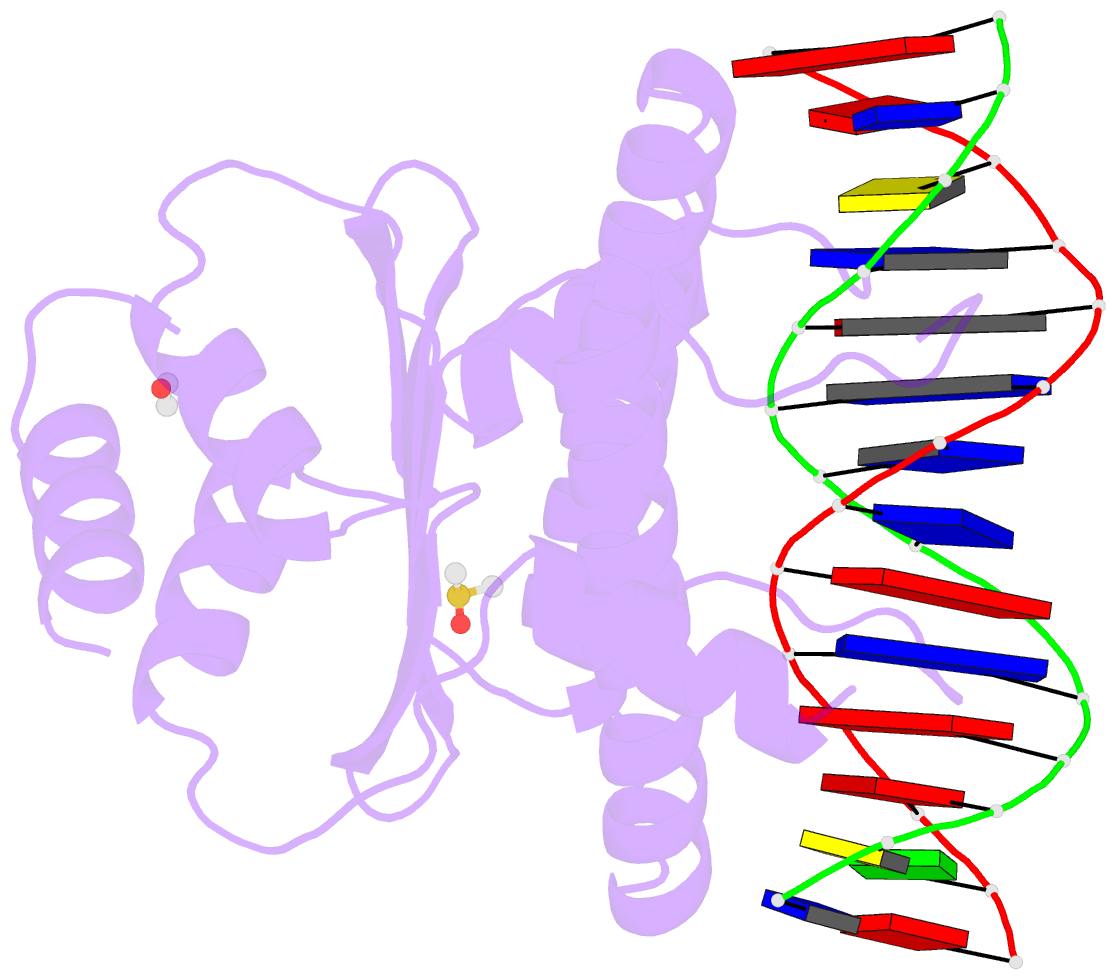
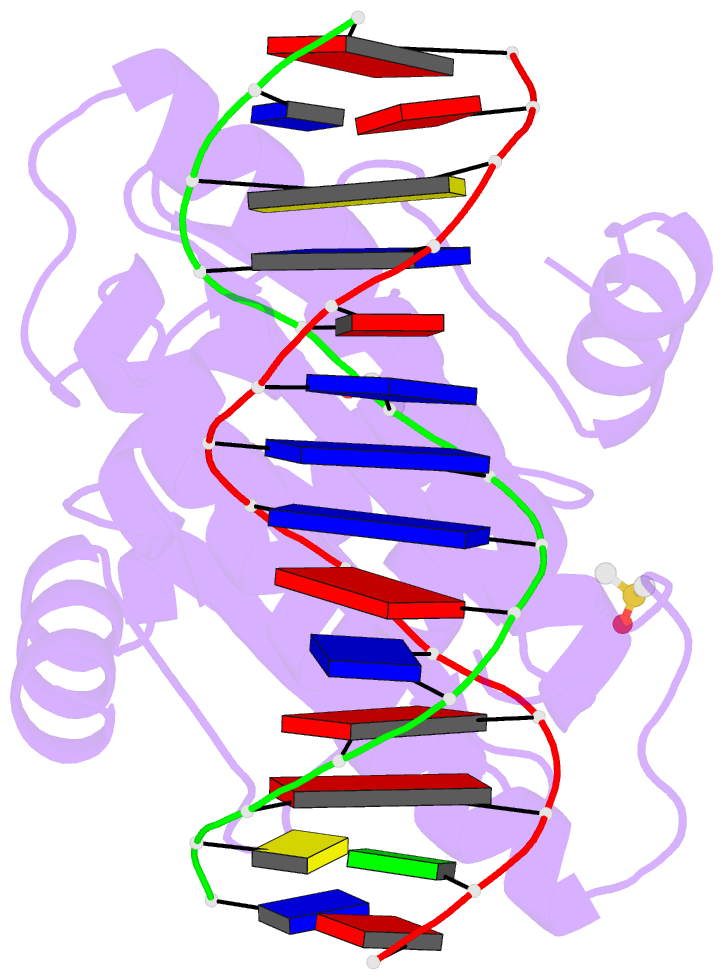
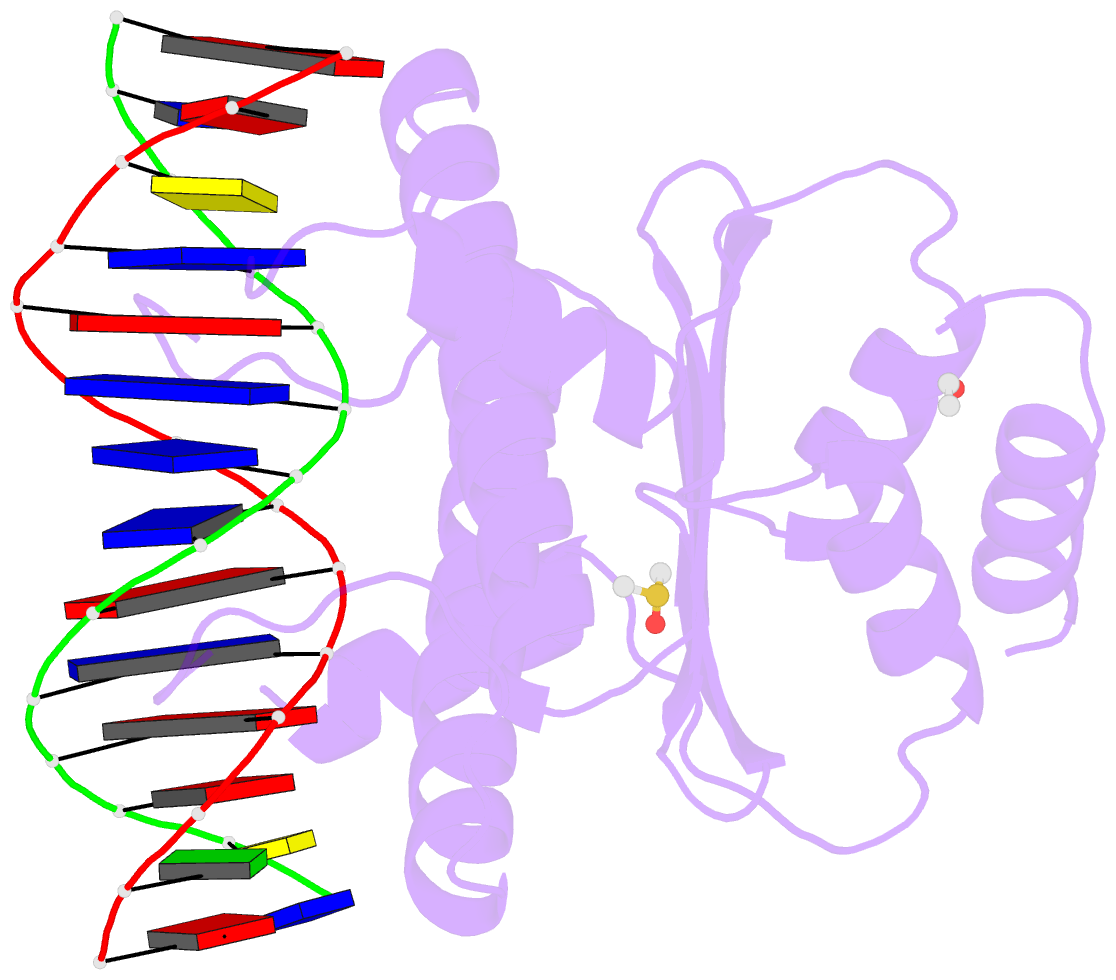
- Summary
- Crystal structure of the mads-box-mef2 domain of mef2d
bound to dsDNA and mitr deacetylase binding motif mutant
l151v.
- Reference
-
Chinellato M, Perin S, Carli A, Lastella L, Biondi B,
Borsato G, Di Giorgio E, Brancolini C, Cendron L,
Angelini A (2024): "Folding of
Class IIa HDAC Derived Peptides into alpha-helices Upon
Binding to Myocyte Enhancer Factor-2 in Complex with
DNA." J.Mol.Biol., 436,
168541. doi: 10.1016/j.jmb.2024.168541.
- Abstract
- Interaction of transcription factor myocyte enhancer
factor-2 (MEF2) family members with class IIa histone
deacetylases (HDACs) has been implicated in a wide variety
of diseases. Though considerable knowledge on this topic
has been accumulated over the years, a high resolution and
detailed analysis of the binding mode of multiple class IIa
HDAC derived peptides with MEF2D is still lacking. To
fulfil this gap, we report here the crystal structure of
MEF2D in complex with double strand DNA and four different
class IIa HDAC derived peptides, namely HDAC4, HDAC5, HDAC7
and HDAC9. All class IIa HDAC derived peptides form
extended amphipathic α-helix structures that fit snugly in
the hydrophobic groove of MEF2D domain. Binding mode of
class IIa HDAC derived peptides to MEF2D is very similar
and occur primarily through nonpolar interactions mediated
by highly conserved branched hydrophobic amino acids.
Further studies revealed that class IIa HDAC derived
peptides are unstructured in solution and appear to adopt a
folded α-helix structure only upon binding to MEF2D.
Comparison of our peptide-protein complexes with previously
characterized structures of MEF2 bound to different
co-activators and co-repressors, highlighted both
differences and similarities, and revealed the adaptability
of MEF2 in protein-protein interactions. The elucidation of
the three-dimensional structure of MEF2D in complex with
multiple class IIa HDAC derived peptides provide not only a
better understanding of the molecular basis of their
interactions but also have implications for the development
of novel antagonist.