Summary information and primary citation
- PDB-id
-
8jsn;
SNAP-derived features in text and
JSON formats
- Class
- viral protein-RNA
- Method
- cryo-EM (3.4 Å)
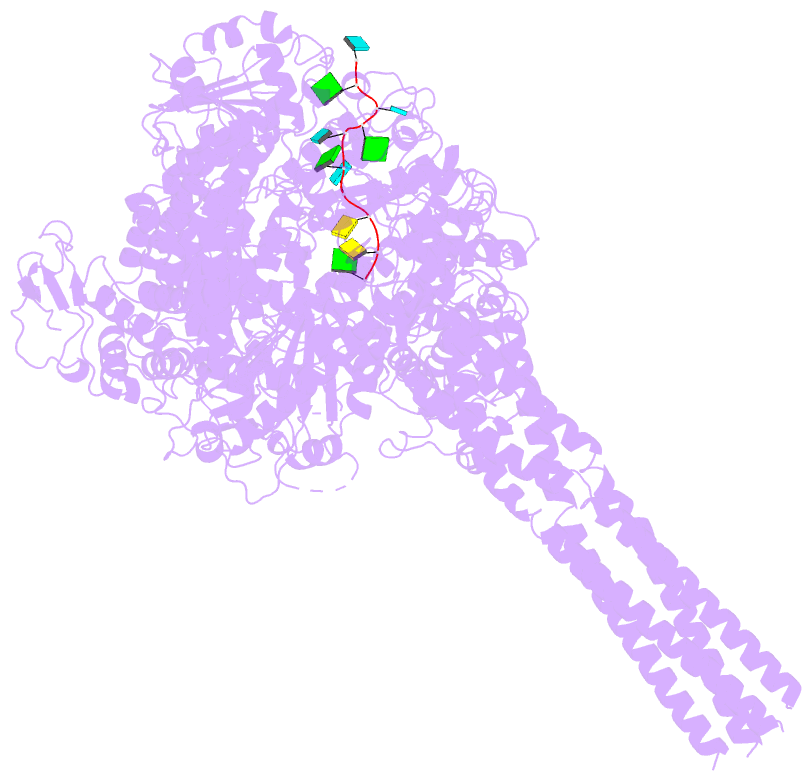
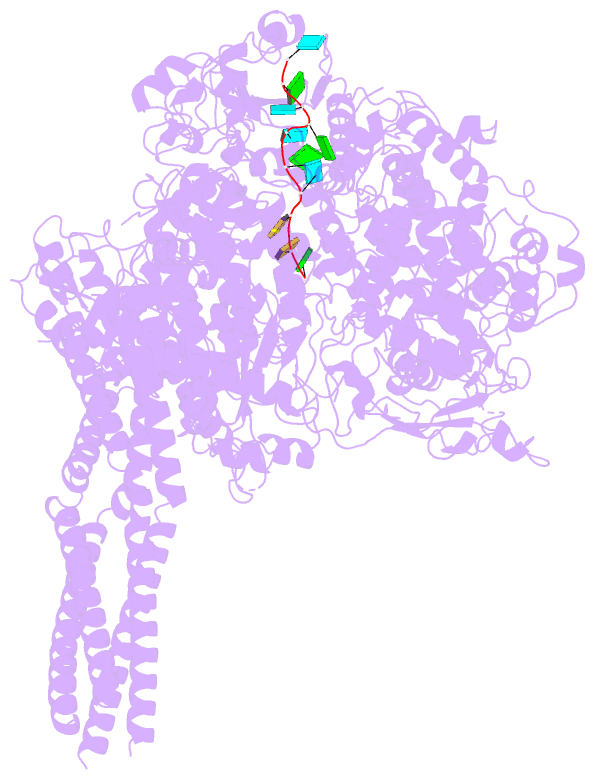
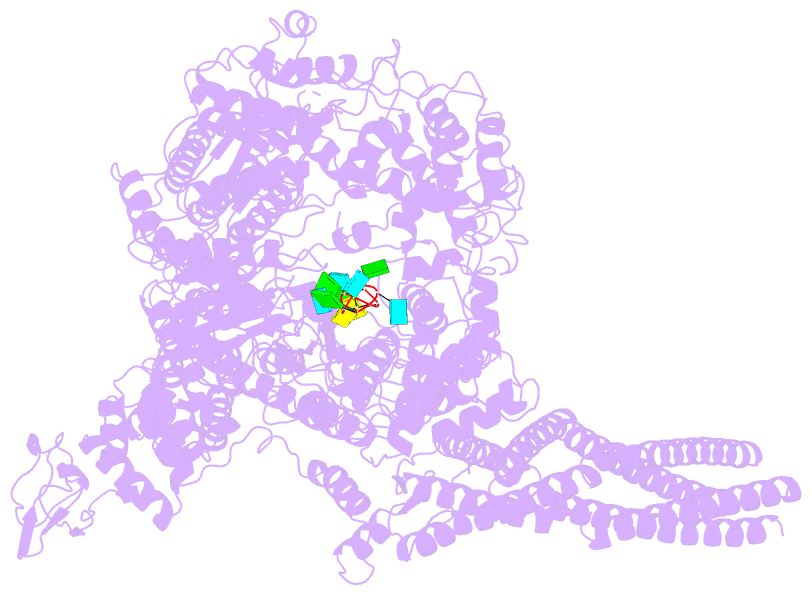
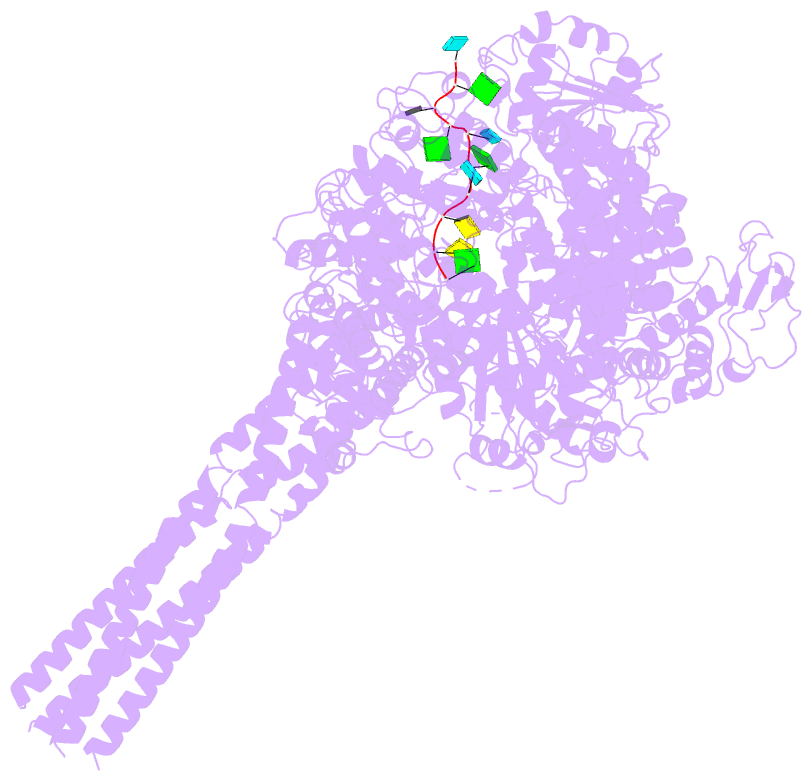
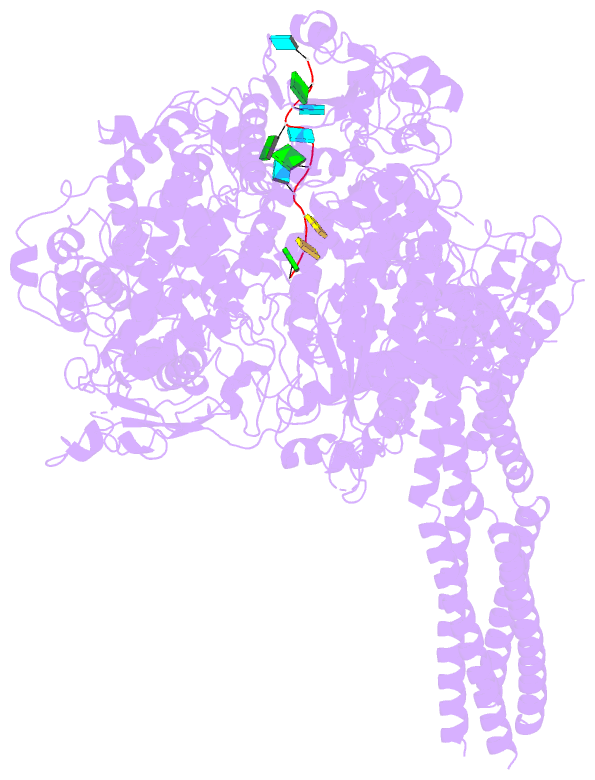
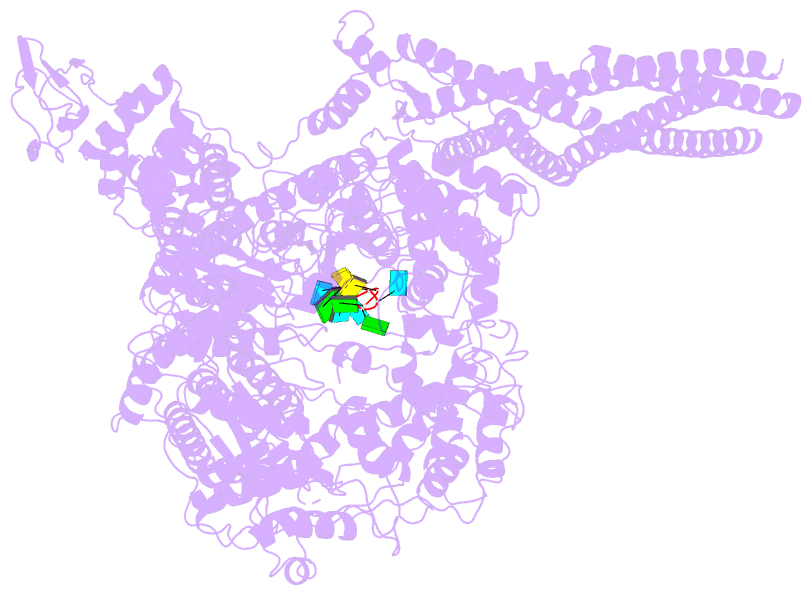
- Summary
- The structure of ebov l-vp35-RNA complex (conformation
2)
- Reference
-
Peng Q, Yuan B, Cheng J, Wang M, Gao S, Bai S, Zhao X, Qi
J, Gao GF, Shi Y (2023): "Molecular
mechanism of de novo replication by the Ebola virus
polymerase." Nature, 622,
603-610. doi: 10.1038/s41586-023-06608-1.
- Abstract
- Non-segmented negative-strand RNA viruses (nsNSVs)
including Ebola virus (EBOV), rabies virus, human
respiratory syncytial virus and pneumoviruses can cause
respiratory infections, hemorrhagic fever and encephalitis
in the humans and animals, and are considered as
substantial health and economic burden
worldwide<sub>1</sub>. Replication and
transcription of the viral genome are executed by the large
L polymerase that is a promising target for the development
of antiviral drugs. Here, using EBOV L polymerase as a
representative, we showed that de novo replication of L
polymerase is controlled by the specific 3' leader sequence
of EBOV genome in an enzymatic assay, and formation of at
least three base pairs can effectively drive the elongation
process of RNA synthesis independent of the specific RNA
sequence. We then determined the high-resolution structures
of EBOV L-VP35-RNA complex and found that the 3' leader RNA
binds in the template entry channel with a distinctive
stable bend conformation. Further mutagenesis work
confirmed that the bend conformation of RNA is required for
the de novo replication activity and revealed the key
residues of L protein that stabilize the RNA conformation.
These findings have provided a new mechanistic
understanding of RNA synthesis for nsNSV polymerases, and
revealed important targets for the development of antiviral
drugs.