Summary information and primary citation
- PDB-id
- 8f2y; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (1.78 Å)
- Summary
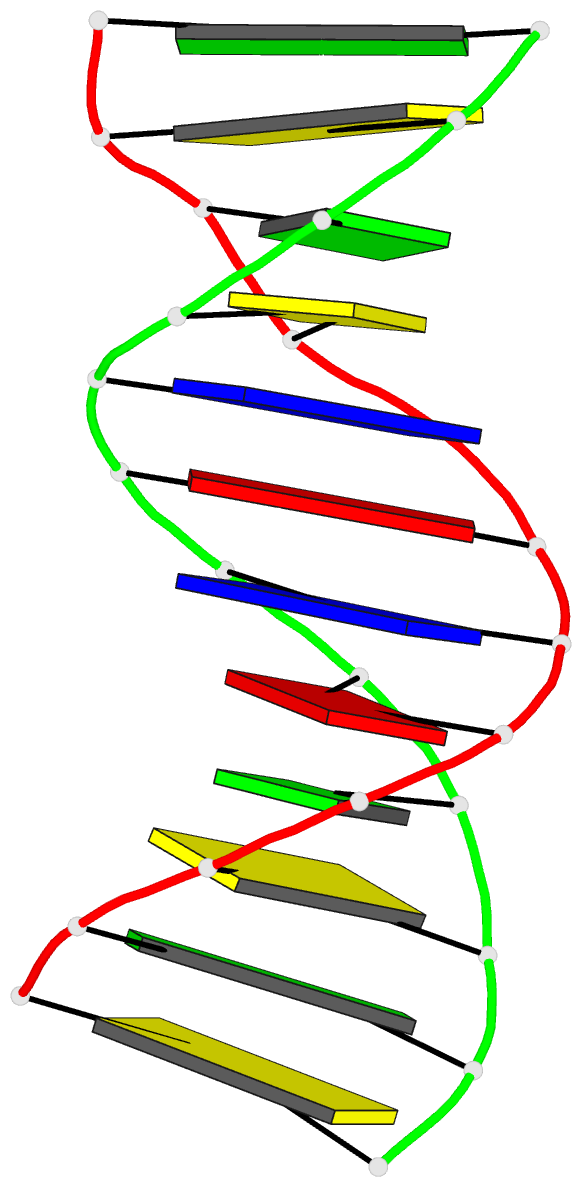
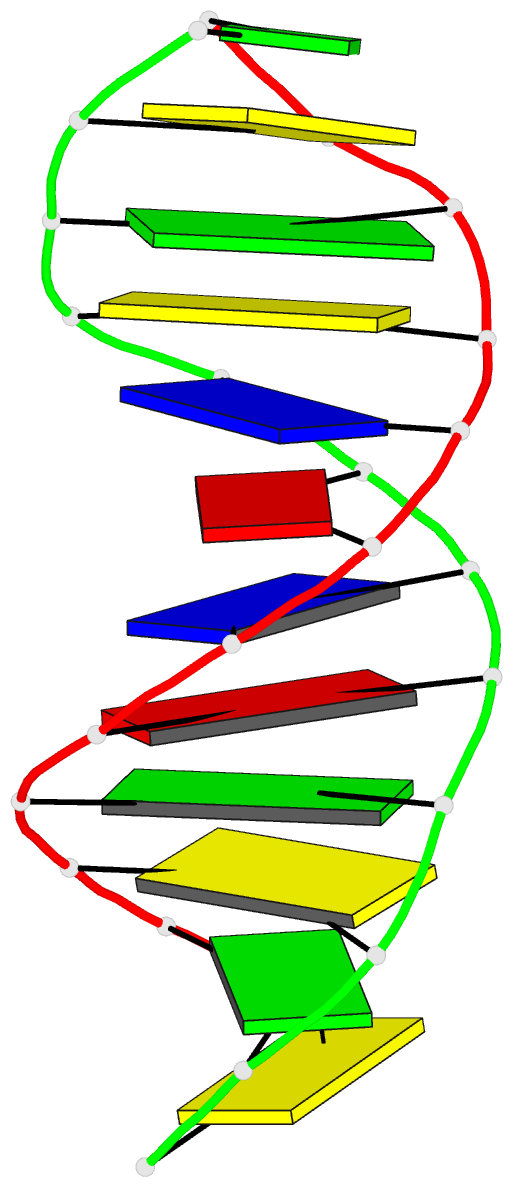
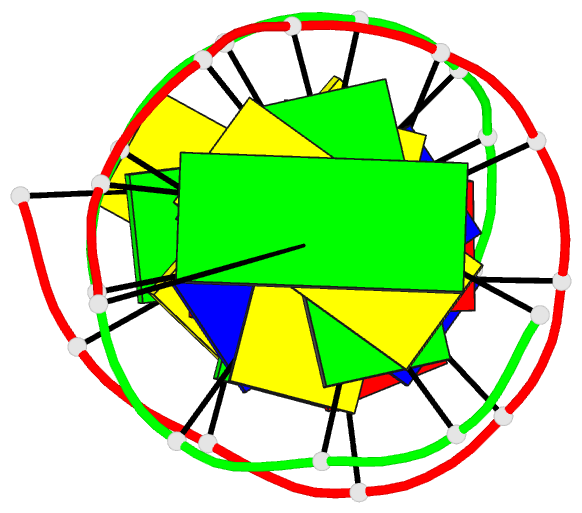
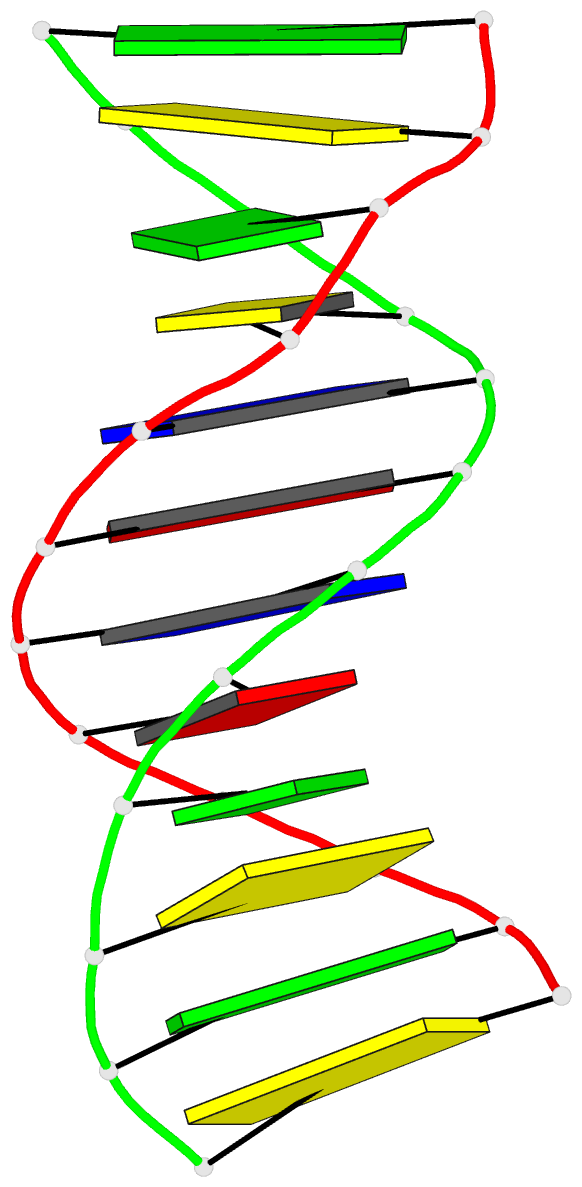
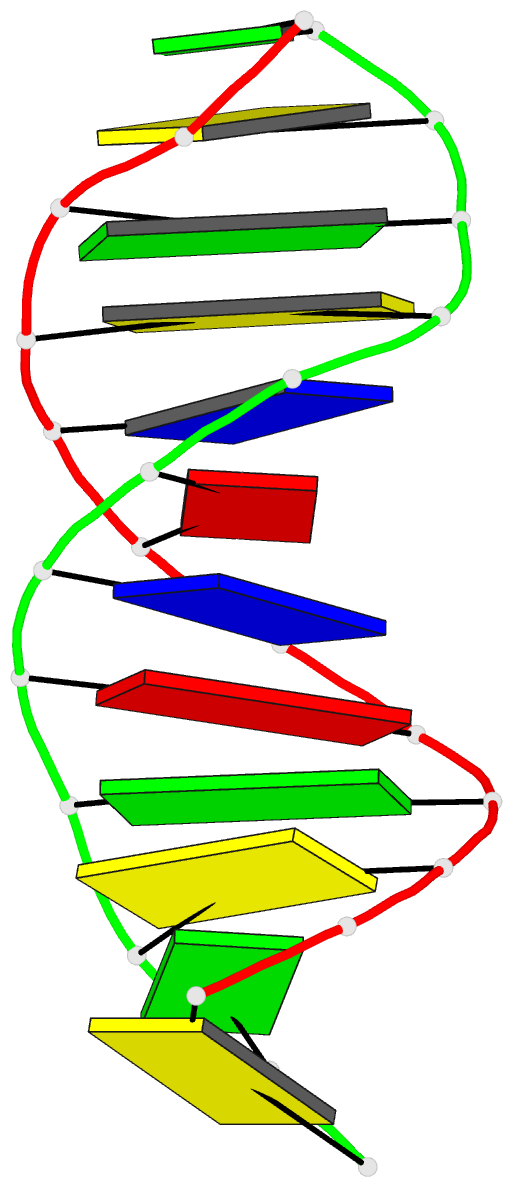
- Structure of an alternating at dodecamer: 5'-cgcgatatcgcg-3
- Reference
- Ogbonna EN, Paul A, Farahat AA, Terrell JR, Mineva E, Ogbonna V, Boykin DW, Wilson WD (2023): "X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences." Acs Bio Med Chem Au, 3, 335-348. doi: 10.1021/acsbiomedchemau.3c00002.
- Abstract
- The rational design of small molecules that target specific DNA sequences is a promising strategy to modulate gene expression. This report focuses on a diamidinobenzimidazole compound, whose selective binding to the minor groove of AT DNA sequences holds broad significance in the molecular recognition of AT-rich human promoter sequences. The objective of this study is to provide a more detailed and systematized understanding, at an atomic level, of the molecular recognition mechanism of different AT-specific sequences by a rationally designed minor groove binder. The specialized method of X-ray crystallography was utilized to investigate how the sequence-dependent recognition properties in general, A-tract, and alternating AT sequences affect the binding of diamidinobenzimidazole in the DNA minor groove. While general and A-tract AT sequences give a narrower minor groove, the alternating AT sequences intrinsically have a wider minor groove which typically constricts upon binding. A strong and direct hydrogen bond between the N-H of the benzimidazole and an H-bond acceptor atom in the minor groove is essential for DNA recognition in all sequences described. In addition, the diamidine compound specifically utilizes an interfacial water molecule for its DNA binding. DNA complexes of AATT and AAAAAA recognition sites show that the diamidine compound can bind in two possible orientations with a preference for water-assisted hydrogen bonding at either cationic end. The complex structures of AAATTT, ATAT, ATATAT, and AAAA are bound in a singular orientation. Analysis of the helical parameters shows a minor groove expansion of about 1 Å across all the nonalternating DNA complexes. The results from this systematic approach will convey a greater understanding of the specific recognition of a diverse array of AT-rich sequences by small molecules and more insight into the design of small molecules with enhanced specificity to AT and mixed DNA sequences.