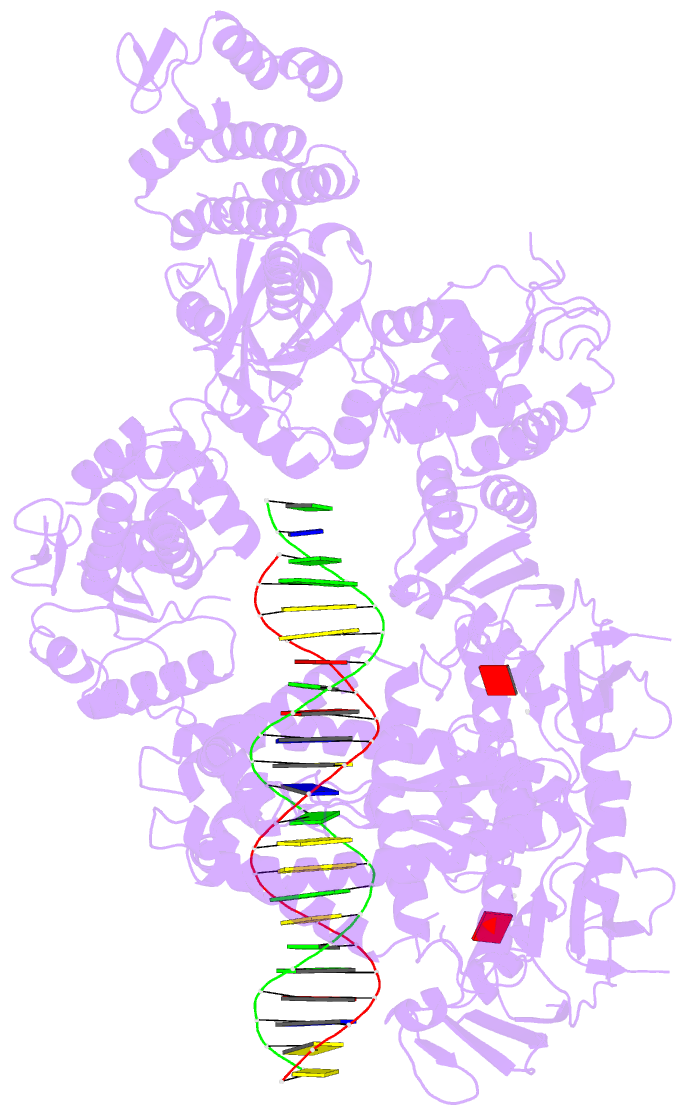
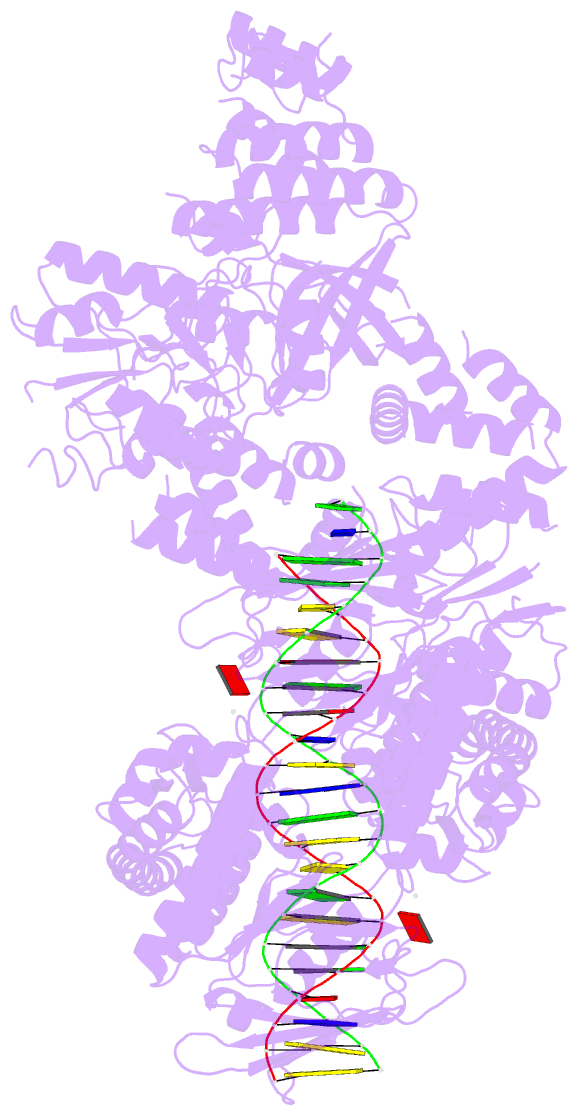
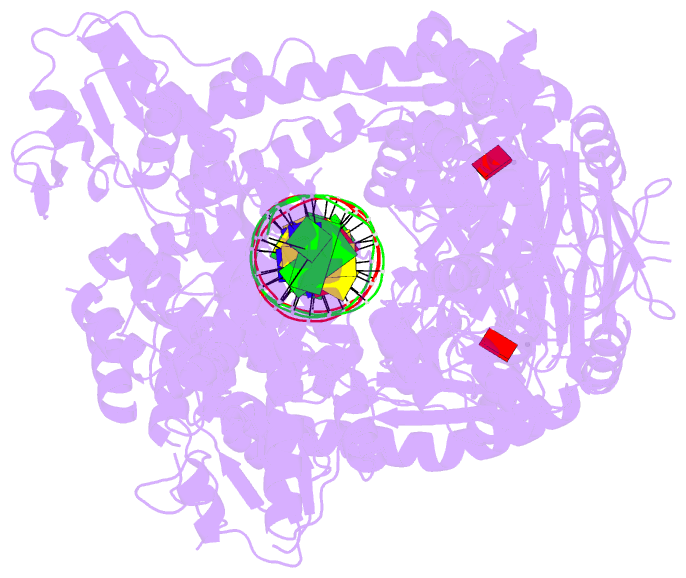
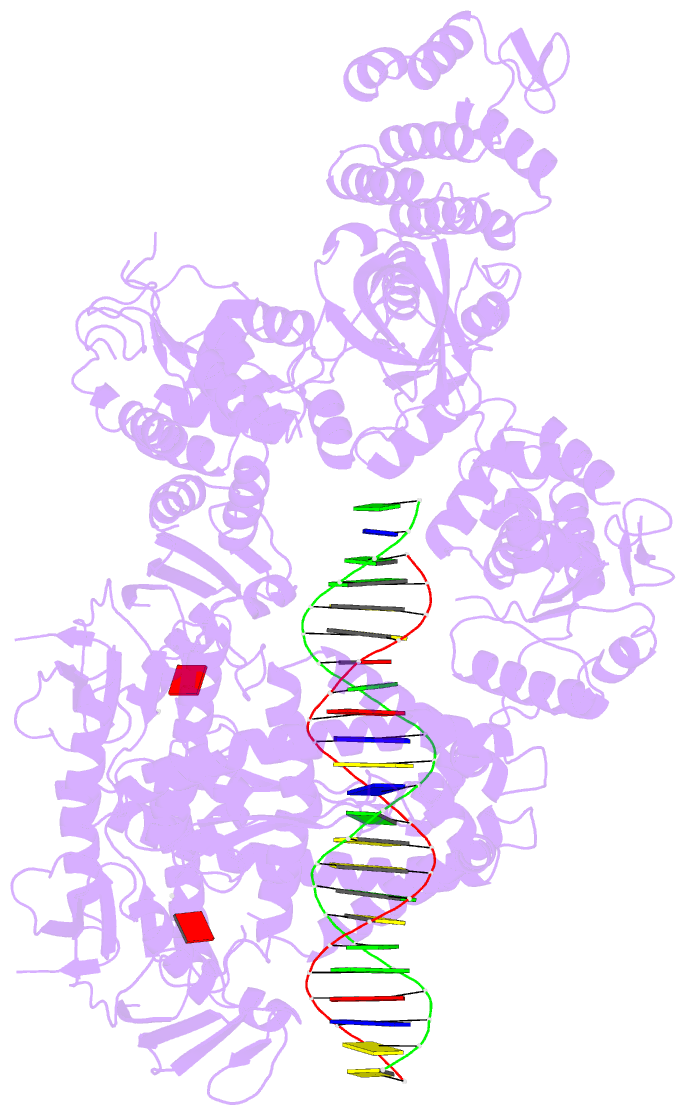
Summary information and primary citation
- PDB-id
- 8bpr; DSSR-derived features in text and JSON formats
- Class
- DNA binding protein
- Method
- cryo-EM (3.65 Å)
- Summary
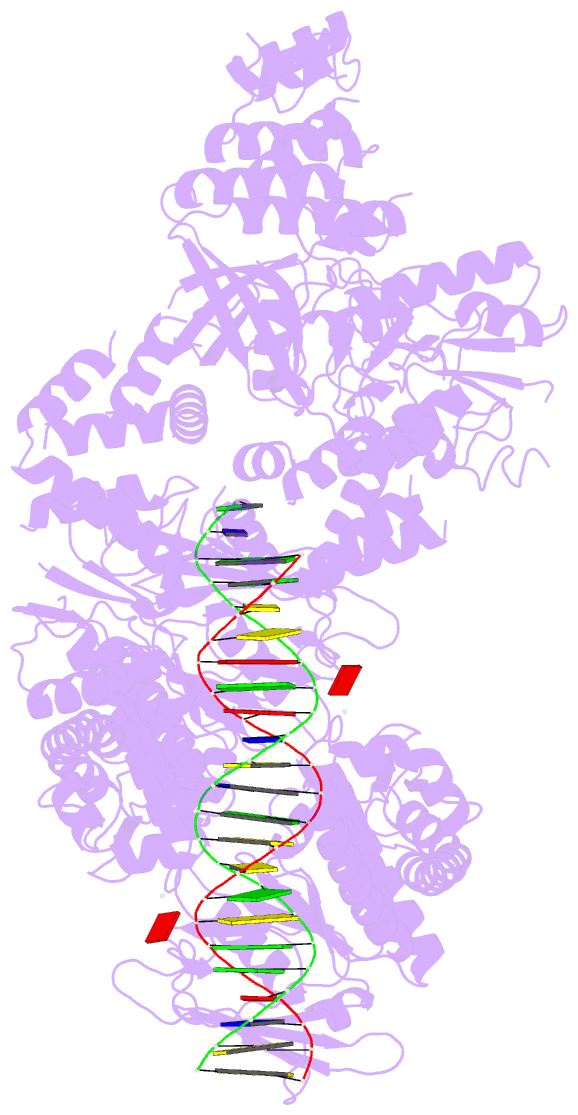
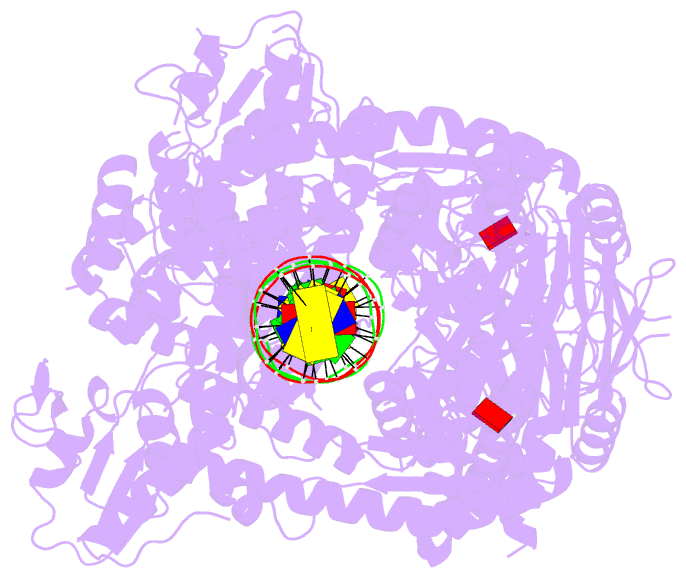
- Complex of recf-reco-recr-DNA from thermus thermophilus (low resolution reconstruction).
- Reference
- Nirwal S, Czarnocki-Cieciura M, Chaudhary A, Zajko W, Skowronek K, Chamera S, Figiel M, Nowotny M (2023): "Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination." Nat.Struct.Mol.Biol., 30, 650-660. doi: 10.1038/s41594-023-00967-z.
- Abstract
- In bacteria, one type of homologous-recombination-based DNA-repair pathway involves RecFOR proteins that bind at the junction between single-stranded (ss) and double-stranded (ds) DNA. They facilitate the replacement of SSB protein, which initially covers ssDNA, with RecA, which mediates the search for homologous sequences. However, the molecular mechanism of RecFOR cooperation remains largely unknown. We used Thermus thermophilus proteins to study this system. Here, we present a cryo-electron microscopy structure of the RecF-dsDNA complex, and another reconstruction that shows how RecF interacts with two different regions of the tetrameric RecR ring. Lower-resolution reconstructions of the RecR-RecO subcomplex and the RecFOR-DNA assembly explain how RecO is positioned to interact with ssDNA and SSB, which is proposed to lock the complex on a ssDNA-dsDNA junction. Our results integrate the biochemical data available for the RecFOR system and provide a framework for its complete understanding.