Summary information and primary citation
- PDB-id
- 7xhn; DSSR-derived features in text and JSON formats
- Class
- cell cycle
- Method
- cryo-EM (3.71 Å)
- Summary
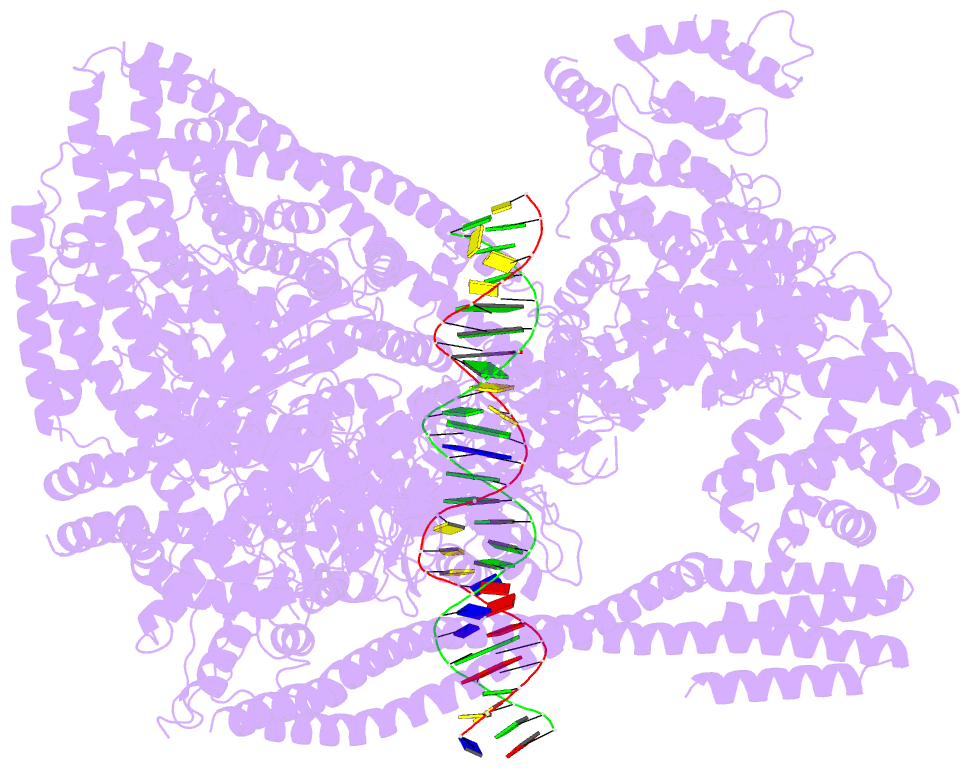
- Structure of human inner kinetochore ccan-DNA complex
- Reference
- Tian T, Chen L, Dou Z, Yang Z, Gao X, Yuan X, Wang C, Liu R, Shen Z, Gui P, Teng M, Meng X, Hill DL, Li L, Zhang X, Liu X, Sun L, Zang J, Yao X (2022): "Structural insights into human CCAN complex assembled onto DNA." Cell Discov, 8, 90. doi: 10.1038/s41421-022-00439-6.
- Abstract
- In mitosis, accurate chromosome segregation depends on kinetochores that connect centromeric chromatin to spindle microtubules. The centromeres of budding yeast, which are relatively simple, are connected to individual microtubules via a kinetochore constitutive centromere associated network (CCAN). However, the complex centromeres of human chromosomes comprise millions of DNA base pairs and attach to multiple microtubules. Here, by use of cryo-electron microscopy and functional analyses, we reveal the molecular basis of how human CCAN interacts with duplex DNA and facilitates accurate chromosome segregation. The overall structure relates to the cooperative interactions and interdependency of the constituent sub-complexes of the CCAN. The duplex DNA is topologically entrapped by human CCAN. Further, CENP-N does not bind to the RG-loop of CENP-A but to DNA in the CCAN complex. The DNA binding activity is essential for CENP-LN localization to centromere and chromosome segregation during mitosis. Thus, these analyses provide new insights into mechanisms of action underlying kinetochore assembly and function in mitosis.