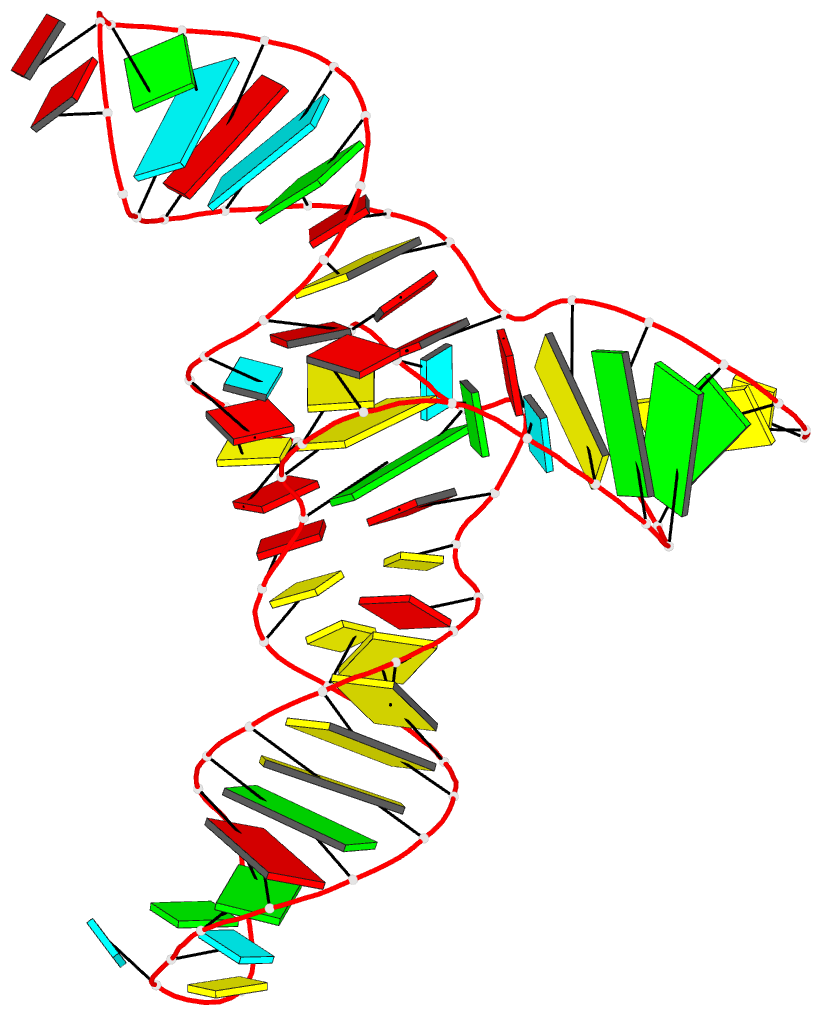
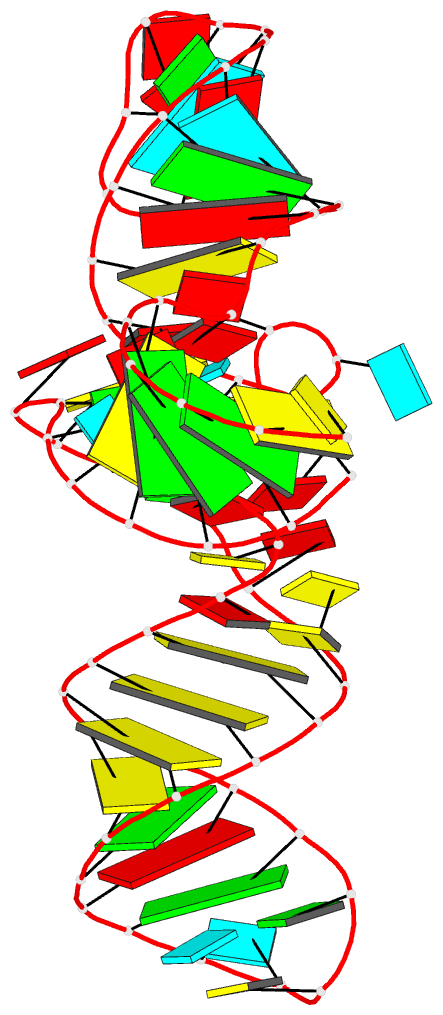
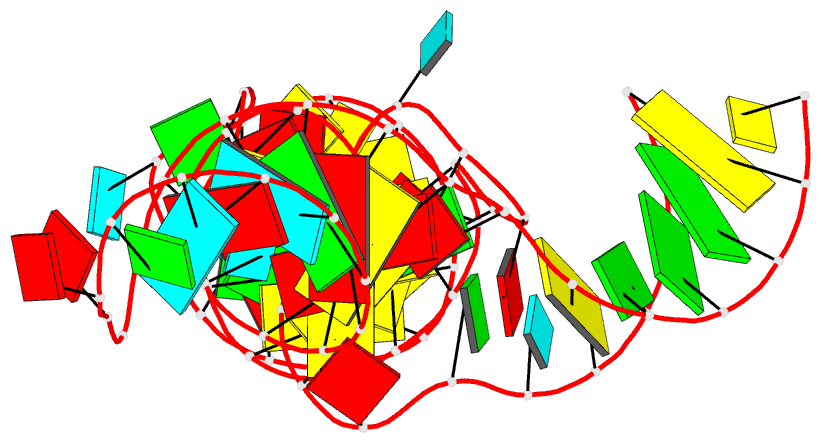
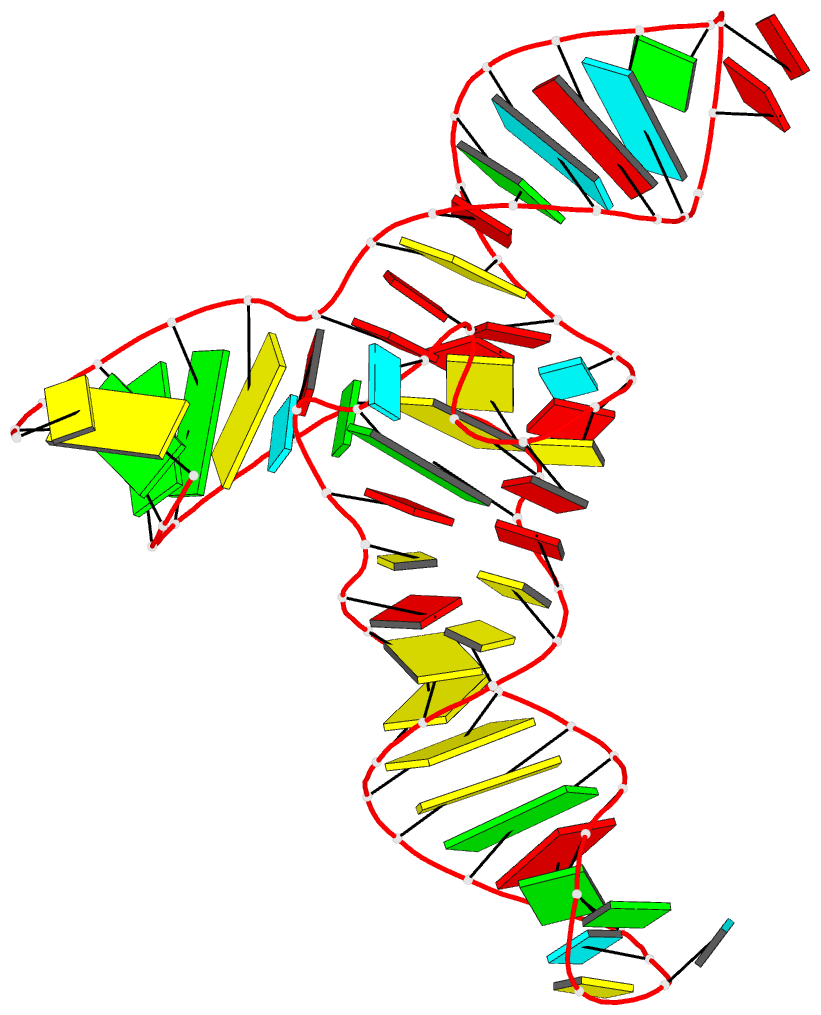
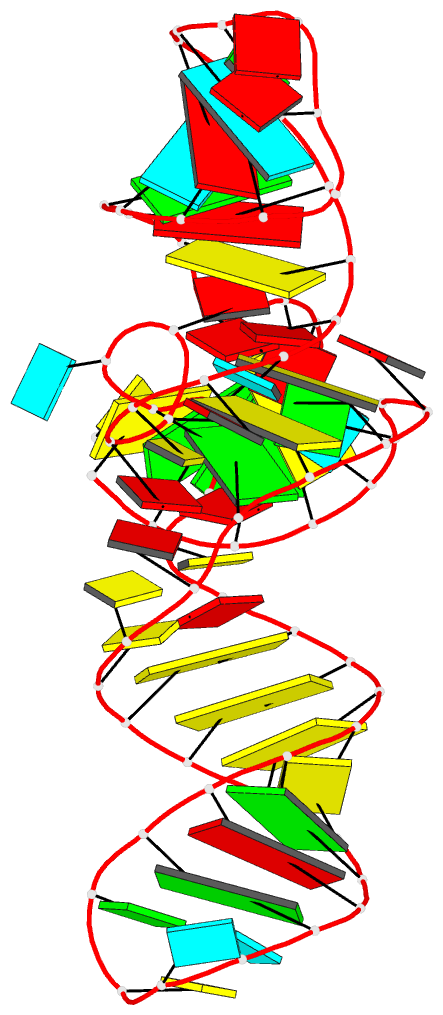
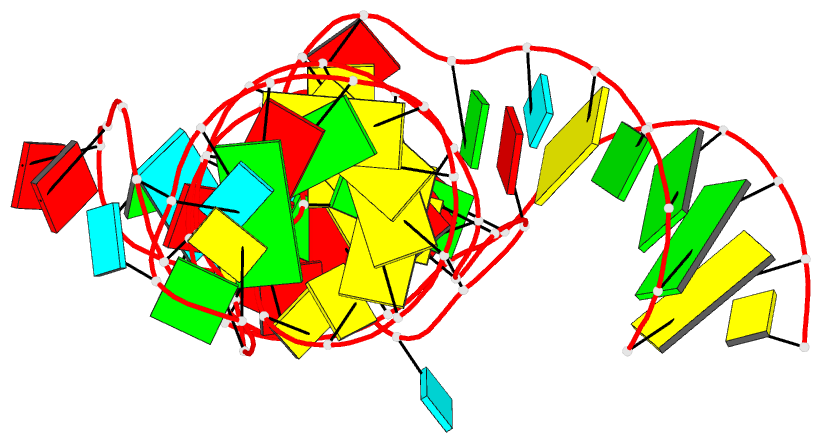
Summary information and primary citation
- PDB-id
- 7v9e; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of a methyl transferase ribozyme
- Reference
- Deng J, Wilson TJ, Wang J, Peng X, Li M, Lin X, Liao W, Lilley DMJ, Huang L (2022): "Structure and mechanism of a methyltransferase ribozyme." Nat.Chem.Biol., 18, 556-564. doi: 10.1038/s41589-022-00982-z.
- Abstract
- Known ribozymes in contemporary biology perform a limited range of chemical catalysis, but in vitro selection has generated species that catalyze a broader range of chemistry; yet, there have been few structural and mechanistic studies of selected ribozymes. A ribozyme has recently been selected that can catalyze a site-specific methyl transfer reaction. We have solved the crystal structure of this ribozyme at a resolution of 2.3 Å, showing how the RNA folds to generate a very specific binding site for the methyl donor substrate. The structure immediately suggests a catalytic mechanism involving a combination of proximity and orientation and nucleobase-mediated general acid catalysis. The mechanism is supported by the pH dependence of the rate of catalysis. A selected methyltransferase ribozyme can thus use a relatively sophisticated catalytic mechanism, broadening the range of known RNA-catalyzed chemistry.