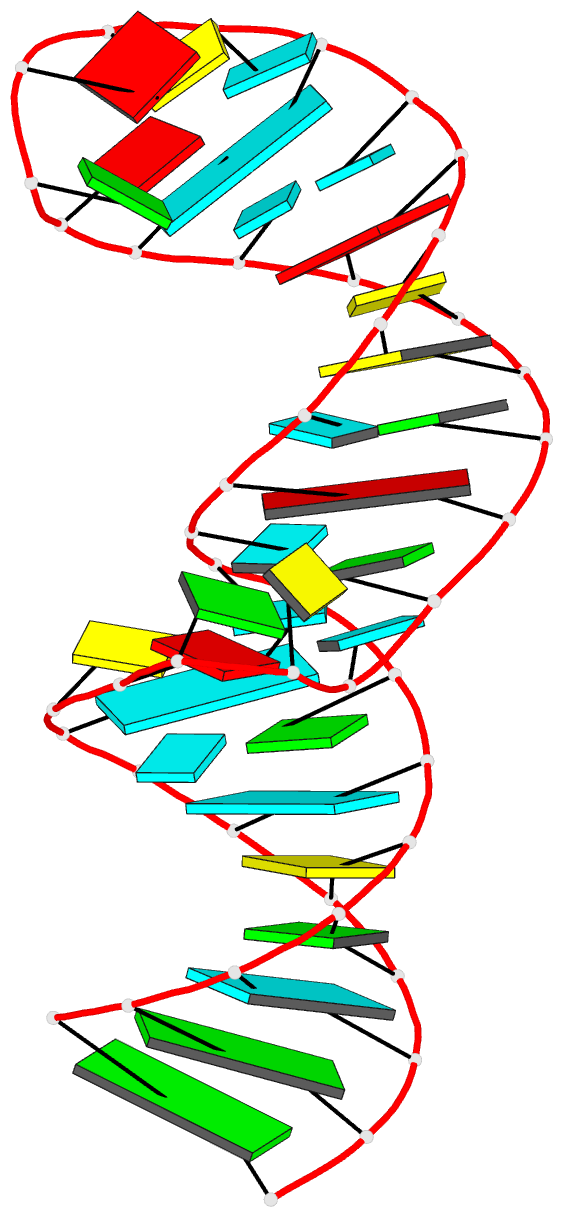
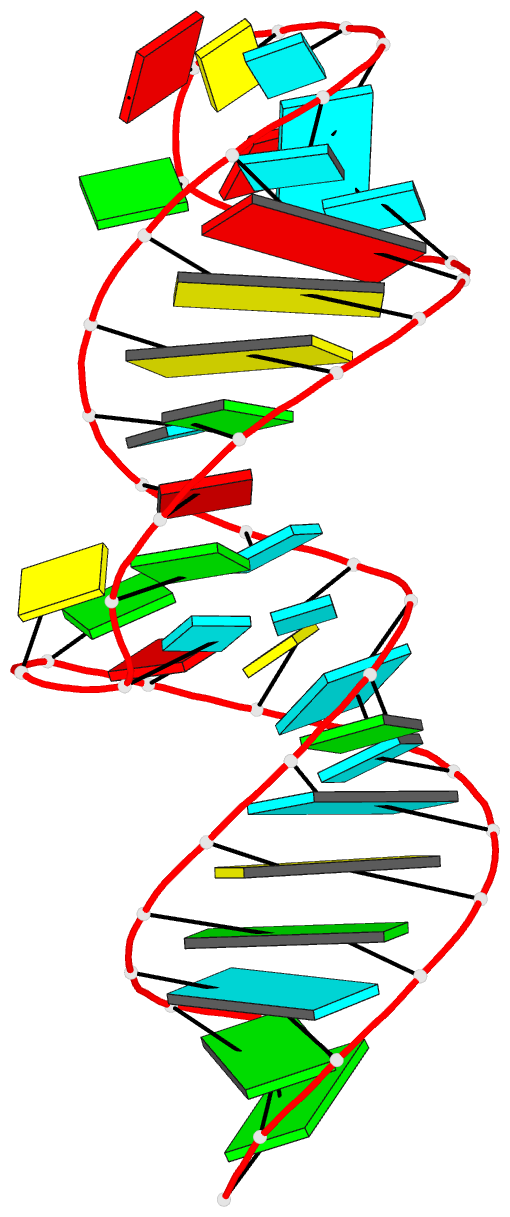
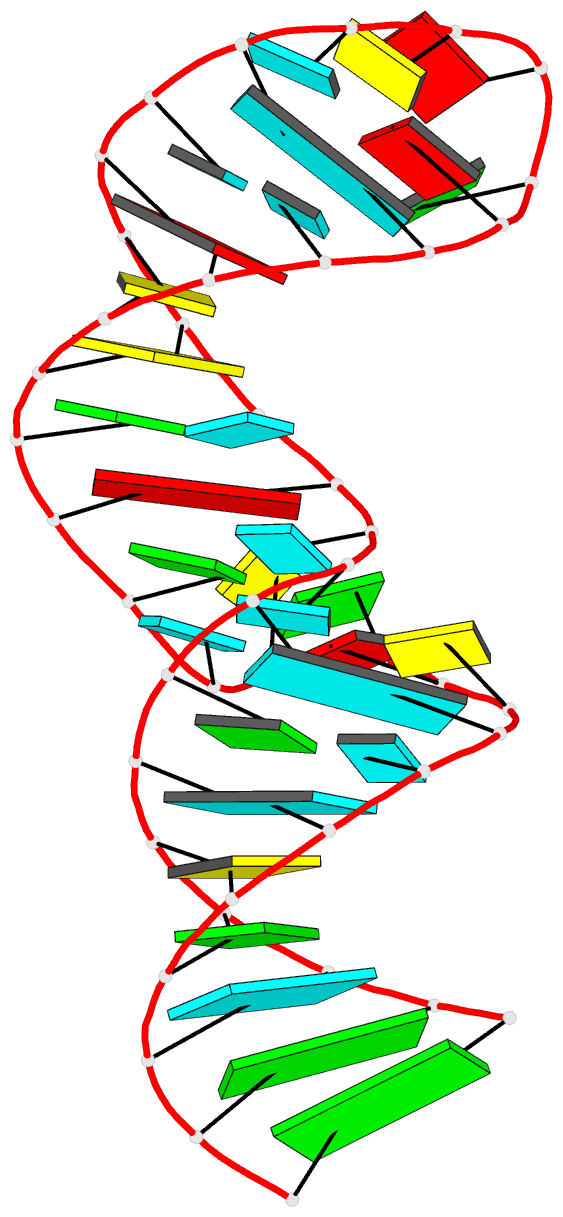
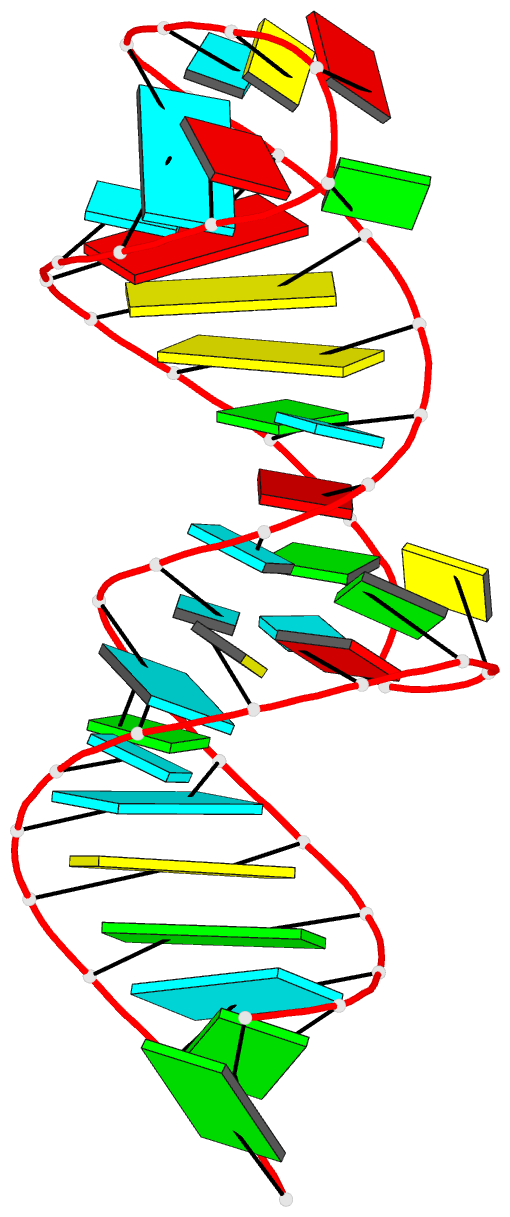
Summary information and primary citation
- PDB-id
- 7v06; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- multiple methods: solution nmr, solution scattering
- Summary
- Encoded conformational dynamics of the hiv splice site a3 regulatory locus: implications for differential binding of hnrnp splicing auxiliary factors
- Reference
- Chiu LY, Emery A, Jain N, Sugarman A, Kendrick N, Luo L, Ford W, Swanstrom R, Tolbert BS (2022): "Encoded Conformational Dynamics of the HIV Splice Site A3 Regulatory Locus: Implications for Differential Binding of hnRNP Splicing Auxiliary Factors." J.Mol.Biol., 167728. doi: 10.1016/j.jmb.2022.167728.
- Abstract
- Alternative splicing of the HIV transcriptome is controlled through cis regulatory elements functioning as enhancers or silencers depending on their context and the type of host RNA binding proteins they recruit. Splice site acceptor A3 (ssA3) is one of the least used acceptor sites in the HIV transcriptome and its activity determines the levels of tat mRNA. Splice acceptor 3 is regulated by a combination of cis regulatory sequences, auxiliary splicing factors, and presumably RNA structure. The mechanisms by which these multiple regulatory components coordinate to determine the frequency in which ssA3 is utilized is poorly understood. By NMR spectroscopy and phylogenetic analysis, we show that the ssA3 regulatory locus is conformationally heterogeneous and that the sequences that encompass the locus are conserved across most HIV isolates. Despite the conformational heterogeneity, the major stem loop (A3SL1) observed in vitro folds to base pair the Polypyrimdine Tract (PPyT) to the Exon Splicing Silencer 2p (ESS2p) element and to a conserved downstream linker. The 3D structure as determined by NMR spectroscopy further reveals that the A3 consensus cleavage site is embedded within a unique stereochemical environment within the apical loop, where it is surrounded by alternating base-base interactions. Despite being described as a receptor for hnRNP H, the ESS2p element is sequestered by base pairing to the 3' end of the PPyT and within this context it cannot form a stable complex with hnRNP H. By comparison, hnRNP A1 directly binds to the A3 consensus cleavage site located within the apical loop, suggesting that it can directly modulate U2AF assembly. Sequence mutations designed to destabilize the PPyT:ESS2p helix results in an increase usage of ssA3 within HIV-infected cells, consistent with the PPyT becoming more accessible for U2AF recognition. Additional mutations introduced into the downstream ESS2 element synergize with ESS2p to cause further increases in ssA3 usage. When taken together, our work provides a unifying picture by which cis regulatory sequences, splicing auxiliary factors and RNA structure cooperate to provide stringent control over ssA3. We describe this as the pair-and-lock mechanism to restrict access of the PPyT, and posit that it operates to regulate a subset of the heterogenous structures encompassing the ssA3 regulatory locus.