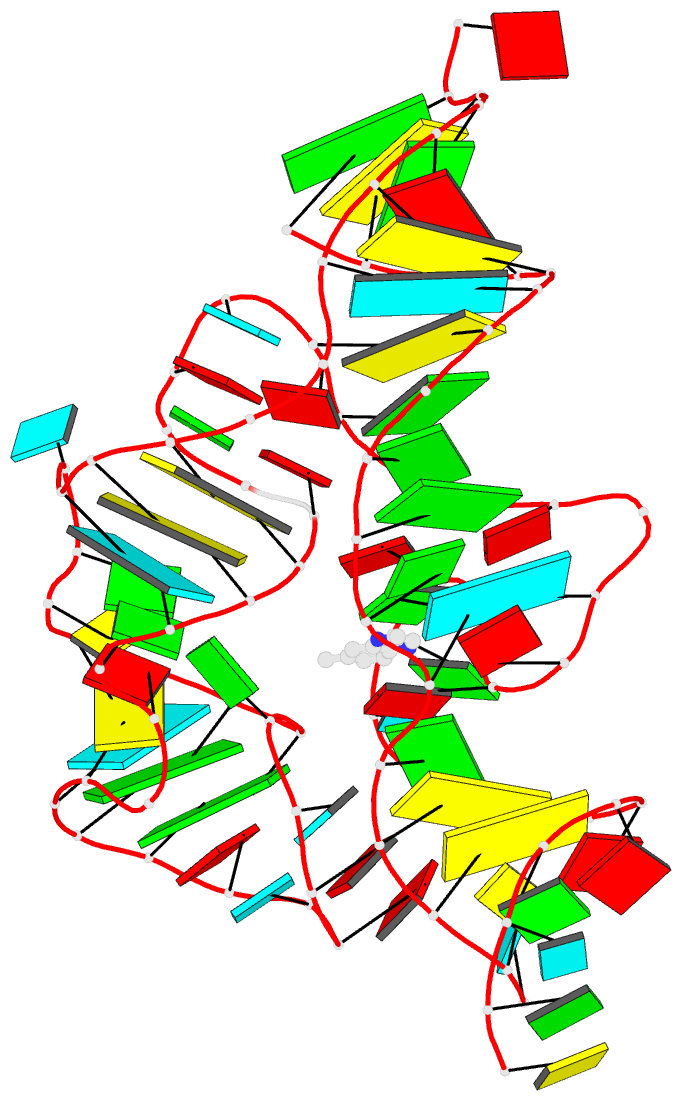
Summary information and primary citation
- PDB-id
- 7tzt; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.96 Å)
- Summary
- Crystal structure of the e. coli thim riboswitch in complex with n1,n1-dimethyl-n2-(quinoxalin-6-ylmethyl)ethane-1,2-diamine (linked compound 37)
- Reference
- Zeller MJ, Favorov O, Li K, Nuthanakanti A, Hussein D, Michaud A, Lafontaine DA, Busan S, Serganov A, Aube J, Weeks KM (2022): "SHAPE-enabled fragment-based ligand discovery for RNA." Proc.Natl.Acad.Sci.USA, 119, e2122660119. doi: 10.1073/pnas.2122660119.
- Abstract
- SignificanceRNA molecules encode proteins and play numerous regulatory roles in cells. Targeting RNA with small molecules, as is routine with proteins, would create broad opportunities for modulating biology and creating new drugs. However, this opportunity has been difficult to realize because creating novel small molecules that bind RNA, especially using modest resources, is challenging. This study integrates two widely used technologies, SHAPE chemical probing of RNA and fragment-based ligand discovery, to craft an innovative strategy for creating small molecules that bind to and modulate the activity of a structured RNA. The anticipated impact is high because the methods are simple, can be implemented in diverse research and discovery contexts, and lead to realistic druglike molecules.