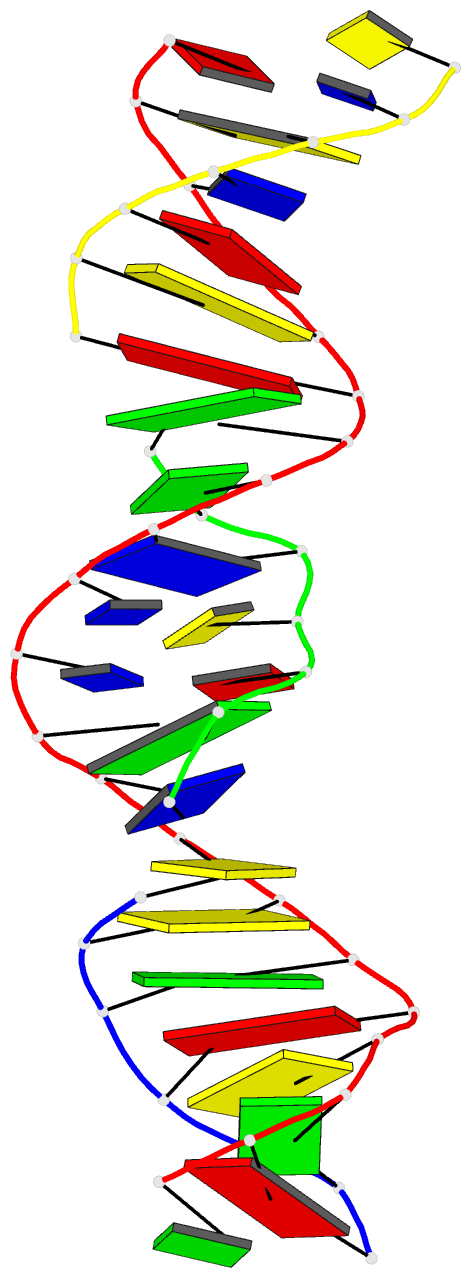
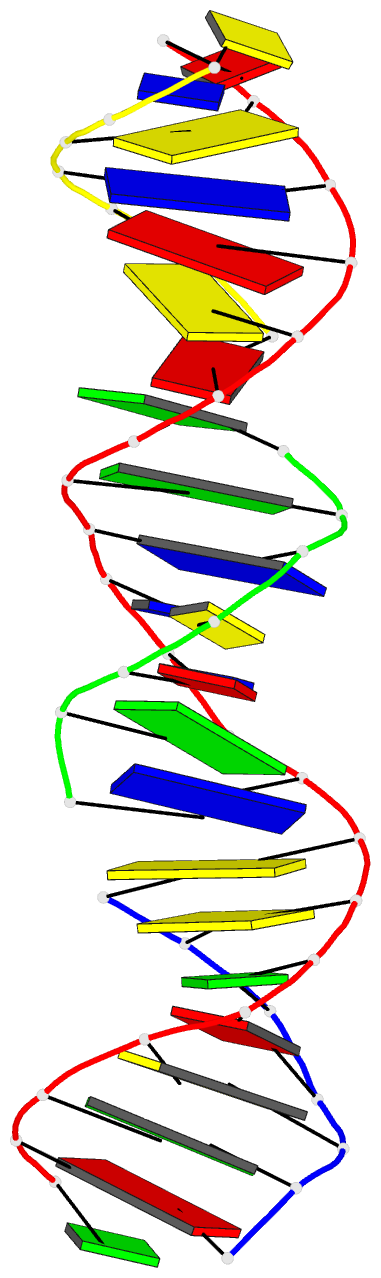
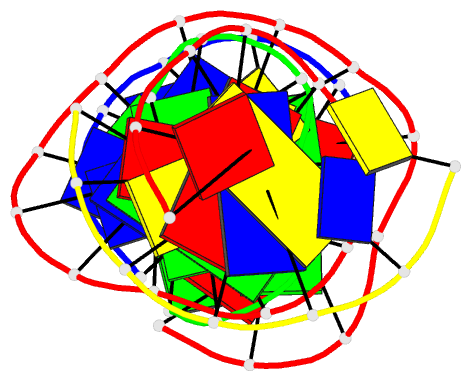
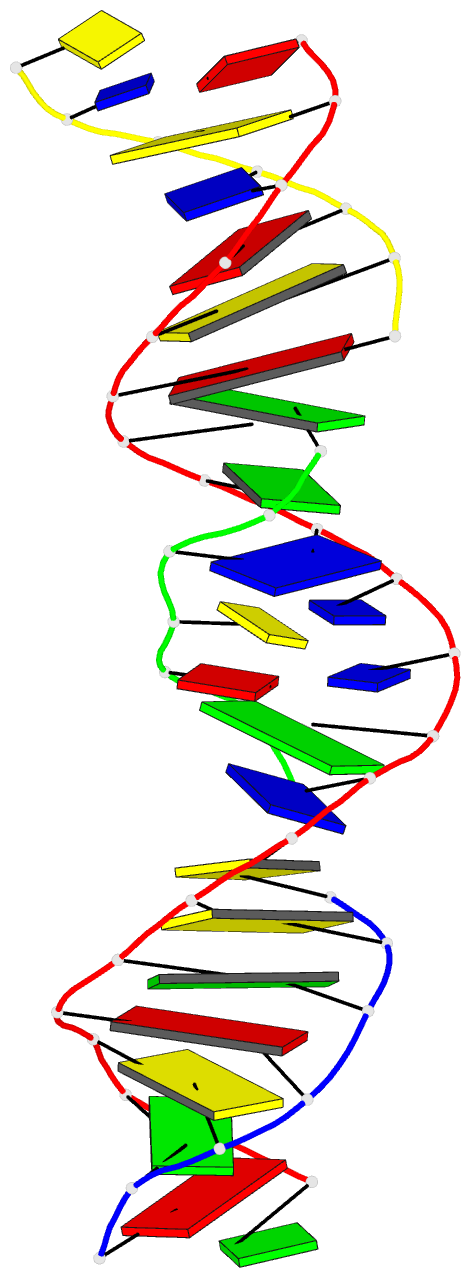
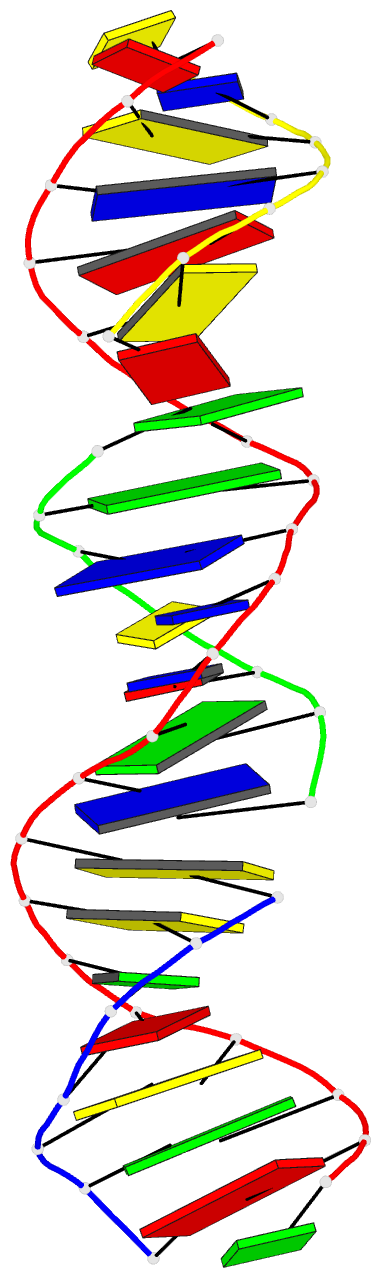
Summary information and primary citation
- PDB-id
- 7sdv; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (4.19 Å)
- Summary
- [t:ag+:mc] metal-mediated DNA base pair in a self-assembling rhombohedral lattice
- Reference
- Vecchioni S, Lu B, Livernois W, Ohayon YP, Yoder JB, Yang CF, Woloszyn K, Bernfeld W, Anantram MP, Canary JW, Hendrickson WA, Rothschild LJ, Mao C, Wind SJ, Seeman NC, Sha R (2023): "Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction." Adv Mater, e2201938. doi: 10.1002/adma.202201938.
- Abstract
- DNA double helices containing metal-mediated DNA (mmDNA) base pairs have been constructed from Ag+ and Hg2+ ions between pyrimidine:pyrimidine pairs with the promise of nanoelectronics. Rational design of mmDNA nanomaterials has been impractical without a complete lexical and structural description. Here, we explore the programmability of structural DNA nanotechnology toward its founding mission of self-assembling a diffraction platform for biomolecular structure determination. We employed the tensegrity triangle to build a comprehensive structural library of mmDNA pairs via X-ray diffraction and elucidated generalized design rules for mmDNA construction. We uncovered two binding modes: N3-dominant, centrosymmetric pairs and major groove binders driven by 5-position ring modifications. Energy gap calculations showed additional levels in the lowest unoccupied molecular orbitals (LUMO) of mmDNA structures, rendering them attractive molecular electronic candidates. This article is protected by copyright. All rights reserved.