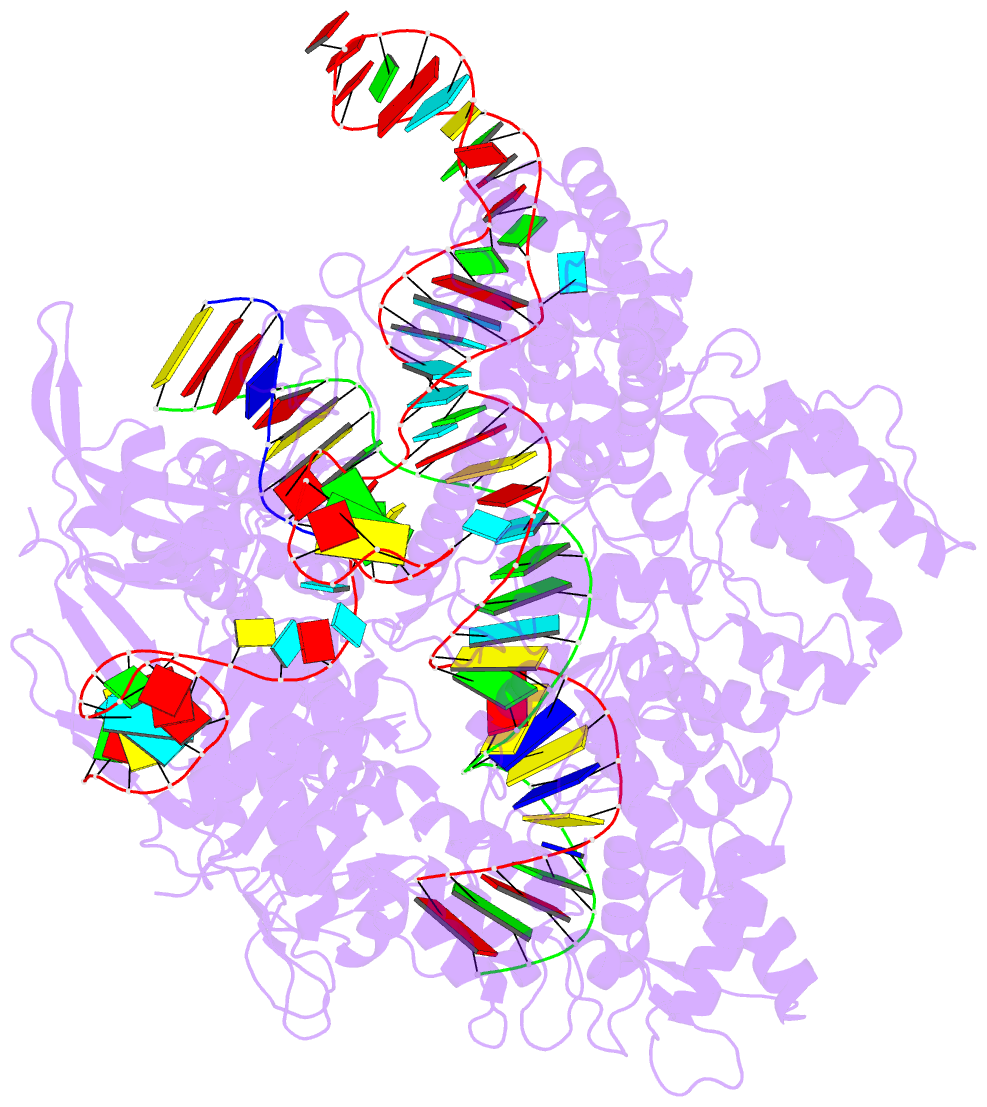
Summary information and primary citation
- PDB-id
- 7ox7; DSSR-derived features in text and JSON formats
- Class
- hydrolase
- Method
- X-ray (2.6 Å)
- Summary
- Target-bound spcas9 complex with trac chimeric RNA-DNA guide
- Reference
- Donohoue PD, Pacesa M, Lau E, Vidal B, Irby MJ, Nyer DB, Rotstein T, Banh L, Toh MS, Gibson J, Kohrs B, Baek K, Owen ALG, Slorach EM, van Overbeek M, Fuller CK, May AP, Jinek M, Cameron P (2021): "Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells." Mol.Cell, 81, 3637-3649.e5. doi: 10.1016/j.molcel.2021.07.035.
- Abstract
- The off-target activity of the CRISPR-associated nuclease Cas9 is a potential concern for therapeutic genome editing applications. Although high-fidelity Cas9 variants have been engineered, they exhibit varying efficiencies and have residual off-target effects, limiting their applicability. Here, we show that CRISPR hybrid RNA-DNA (chRDNA) guides provide an effective approach to increase Cas9 specificity while preserving on-target editing activity. Across multiple genomic targets in primary human T cells, we show that 2'-deoxynucleotide (dnt) positioning affects guide activity and specificity in a target-dependent manner and that this can be used to engineer chRDNA guides with substantially reduced off-target effects. Crystal structures of DNA-bound Cas9-chRDNA complexes reveal distorted guide-target duplex geometry and allosteric modulation of Cas9 conformation. These structural effects increase specificity by perturbing DNA hybridization and modulating Cas9 activation kinetics to disfavor binding and cleavage of off-target substrates. Overall, these results pave the way for utilizing customized chRDNAs in clinical applications.