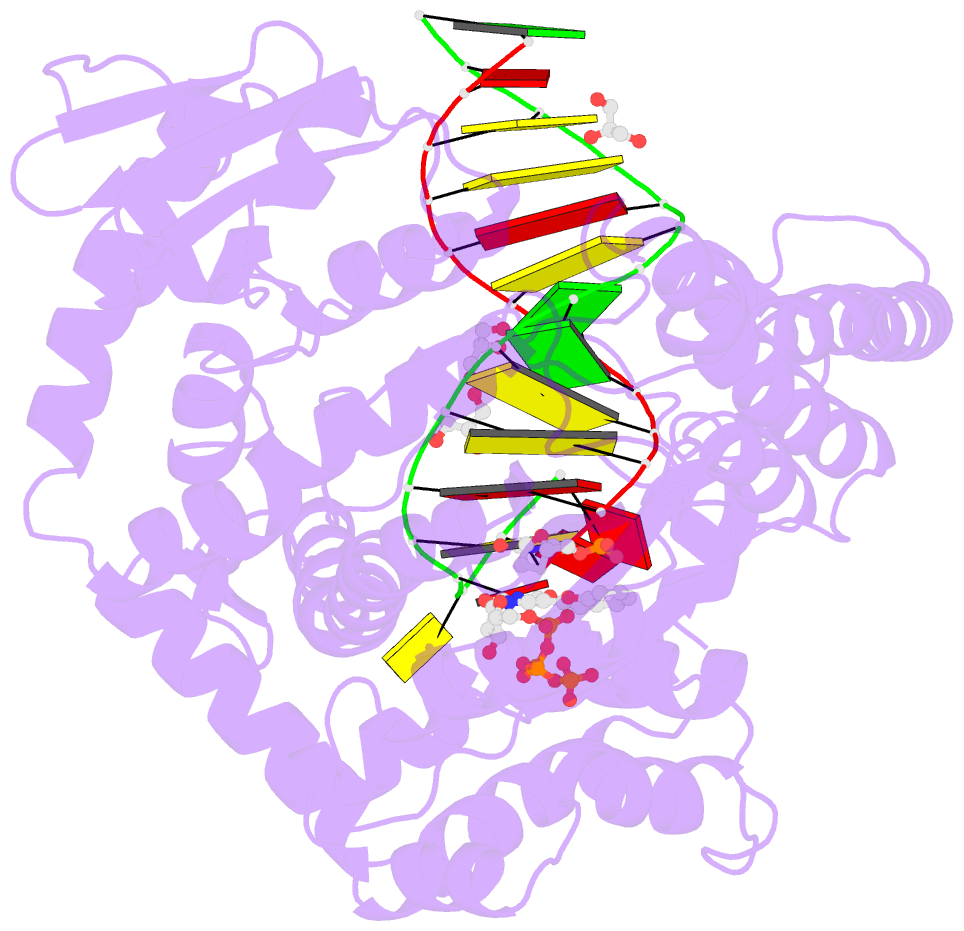
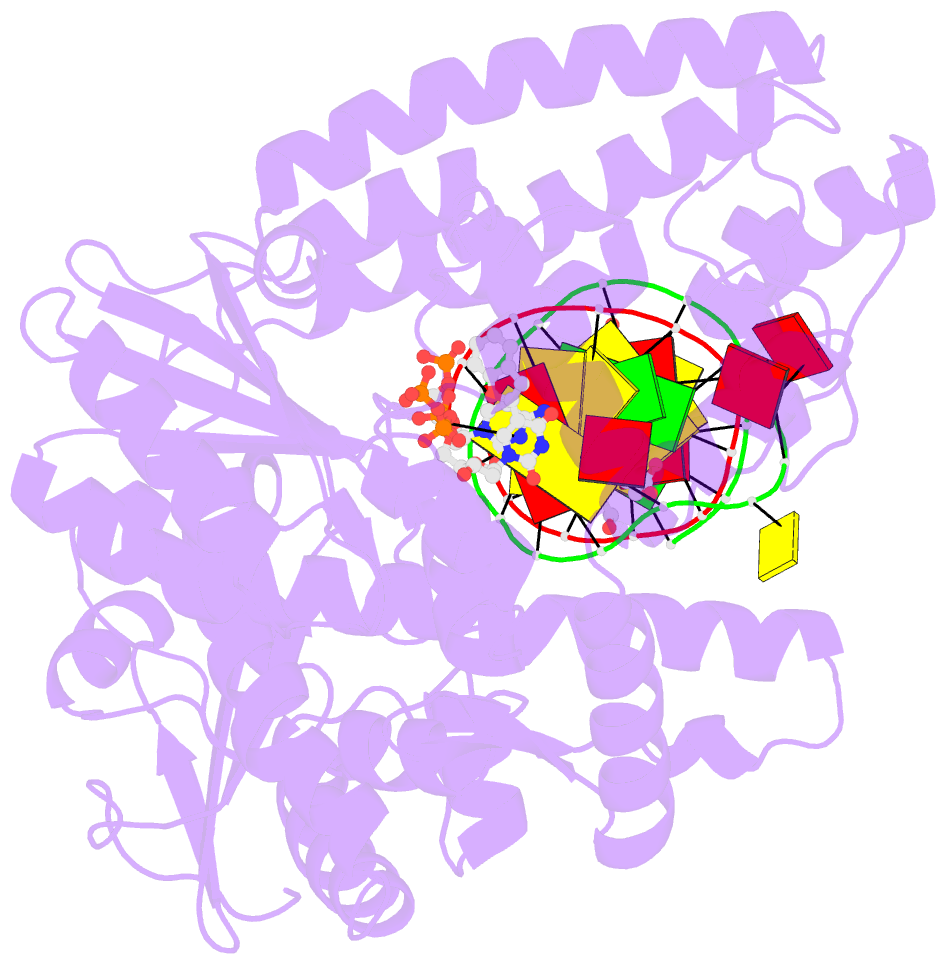
Summary information and primary citation
- PDB-id
- 7owf; DSSR-derived features in text and JSON formats
- Class
- transferase
- Method
- X-ray (1.91 Å)
- Summary
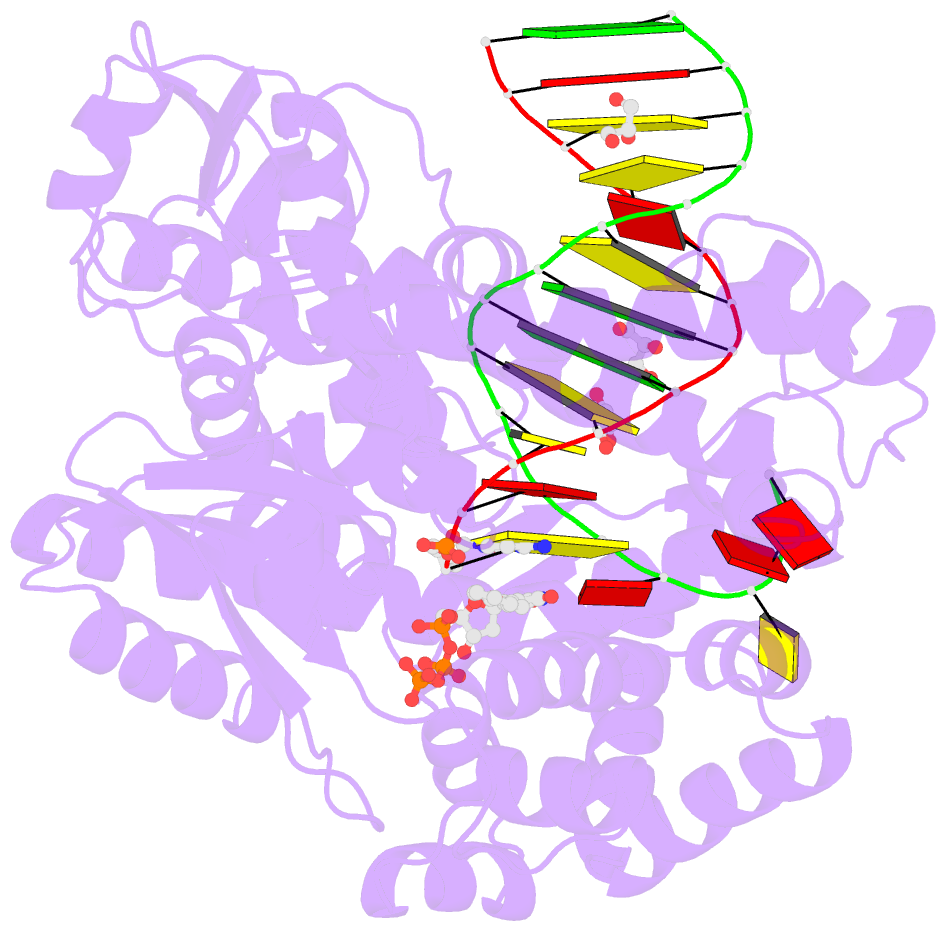
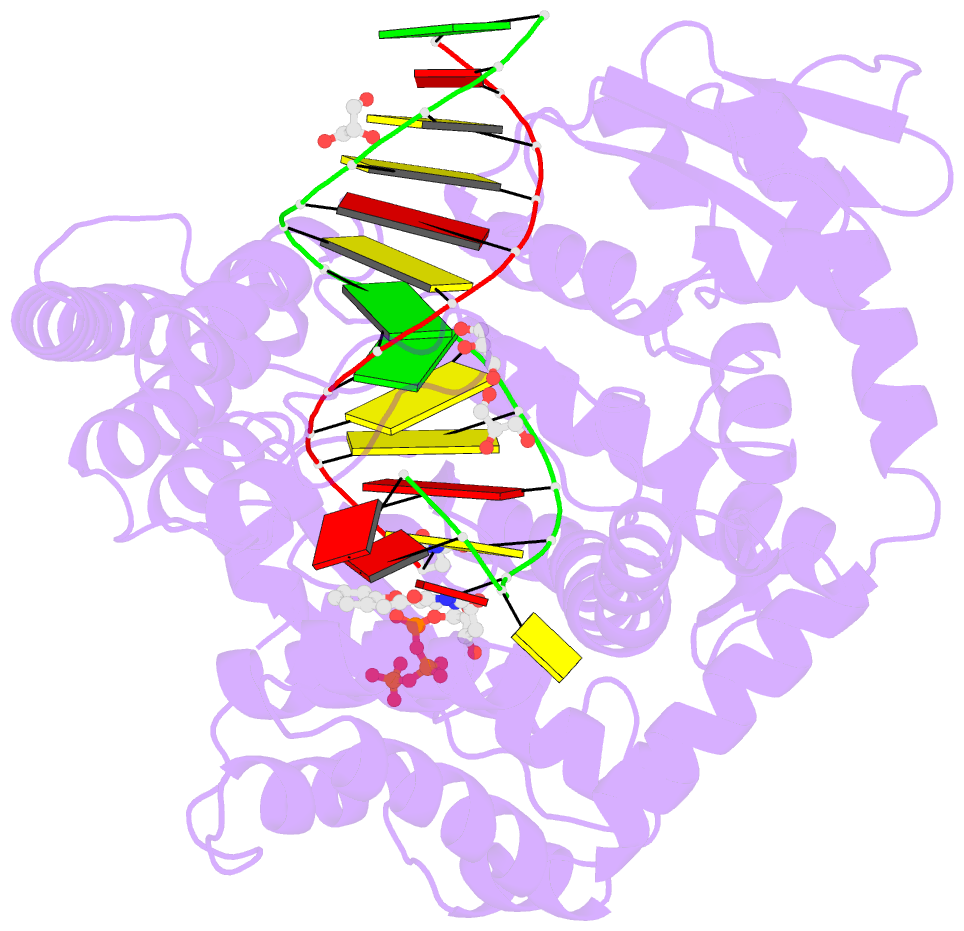
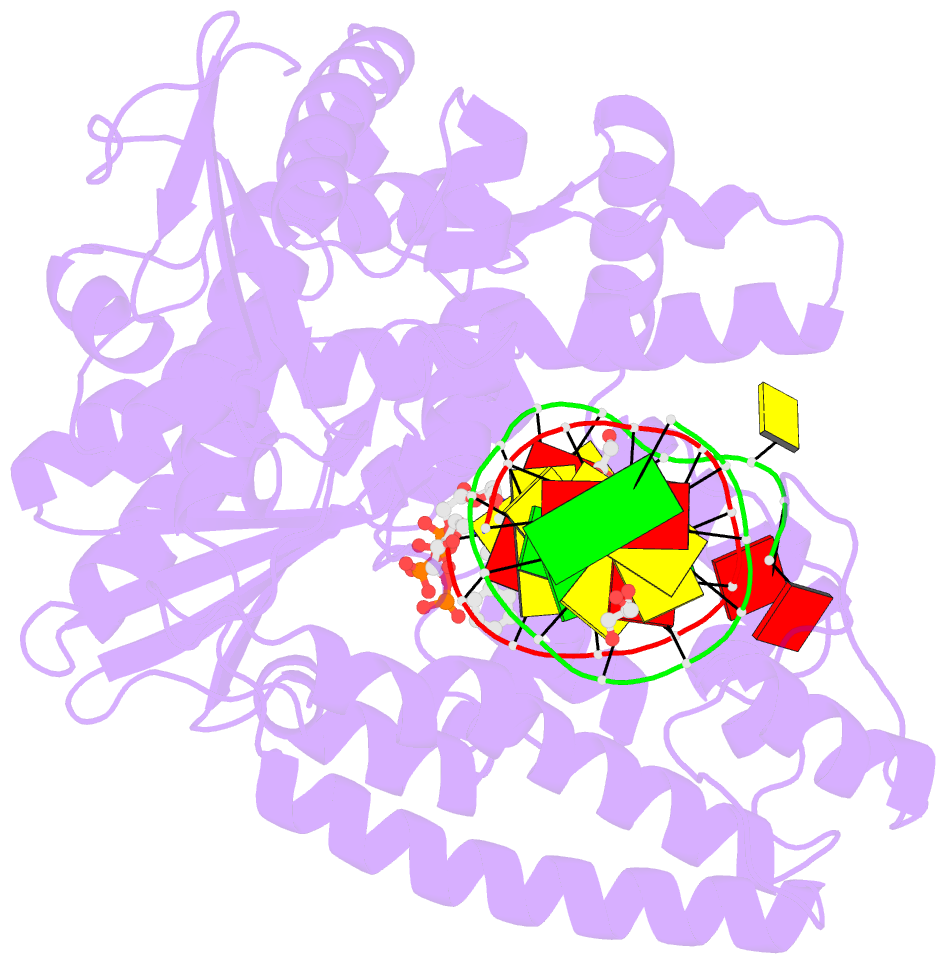
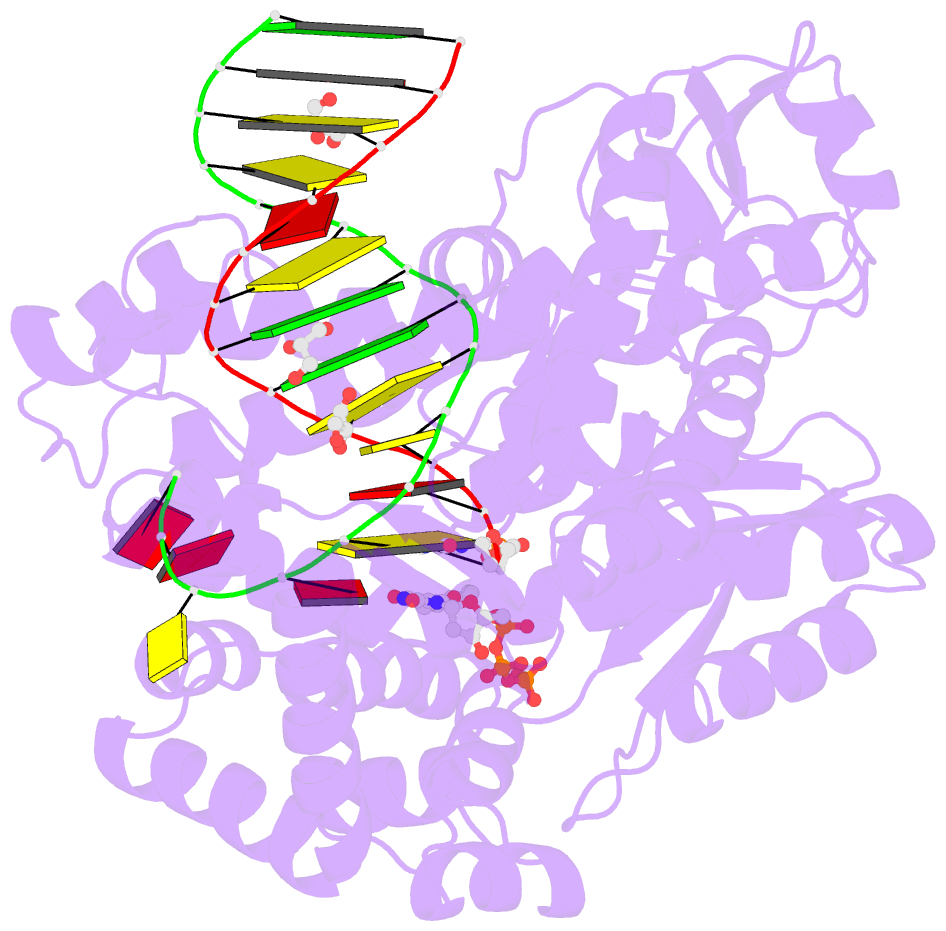
- Klentaq DNA polymerase in a ternary complex with primer-template and the fluorescent nucleotide analog bfdutp
- Reference
- Ghosh P, Kropp HM, Betz K, Ludmann S, Diederichs K, Marx A, Srivatsan SG (2022): "Microenvironment-Sensitive Fluorescent Nucleotide Probes from Benzofuran, Benzothiophene, and Selenophene as Substrates for DNA Polymerases." J.Am.Chem.Soc., 144, 10556-10569. doi: 10.1021/jacs.2c03454.
- Abstract
- DNA polymerases can process a wide variety of structurally diverse nucleotide substrates, but the molecular basis by which the analogs are processed is not completely understood. Here, we demonstrate the utility of environment-sensitive heterocycle-modified fluorescent nucleotide substrates in probing the incorporation mechanism of DNA polymerases in real time and at the atomic level. The nucleotide analogs containing a selenophene, benzofuran, or benzothiophene moiety at the C5 position of 2'-deoxyuridine are incorporated into oligonucleotides (ONs) with varying efficiency, which depends on the size of the heterocycle modification and the DNA polymerase sequence family used. KlenTaq (A family DNA polymerase) is sensitive to the size of the modification as it incorporates only one heterobicycle-modified nucleotide into the growing polymer, whereas it efficiently incorporates the selenophene-modified nucleotide analog at multiple positions. Notably, in the single nucleotide incorporation assay, irrespective of the heterocycle size, it exclusively adds a single nucleotide at the 3'-end of a primer, which enabled devising a simple two-step site-specific ON labeling technique. KOD and Vent(exo-) DNA polymerases, belonging to the B family, tolerate all the three modified nucleotides and produce ONs with multiple labels. Importantly, the benzofuran-modified nucleotide (BFdUTP) serves as an excellent reporter by providing real-time fluorescence readouts to monitor enzyme activity and estimate the binding events in the catalytic cycle. Further, a direct comparison of the incorporation profiles, fluorescence data, and crystal structure of a ternary complex of KlenTaq DNA polymerase with BFdUTP poised for catalysis provides a detailed understanding of the mechanism of incorporation of heterocycle-modified nucleotides.