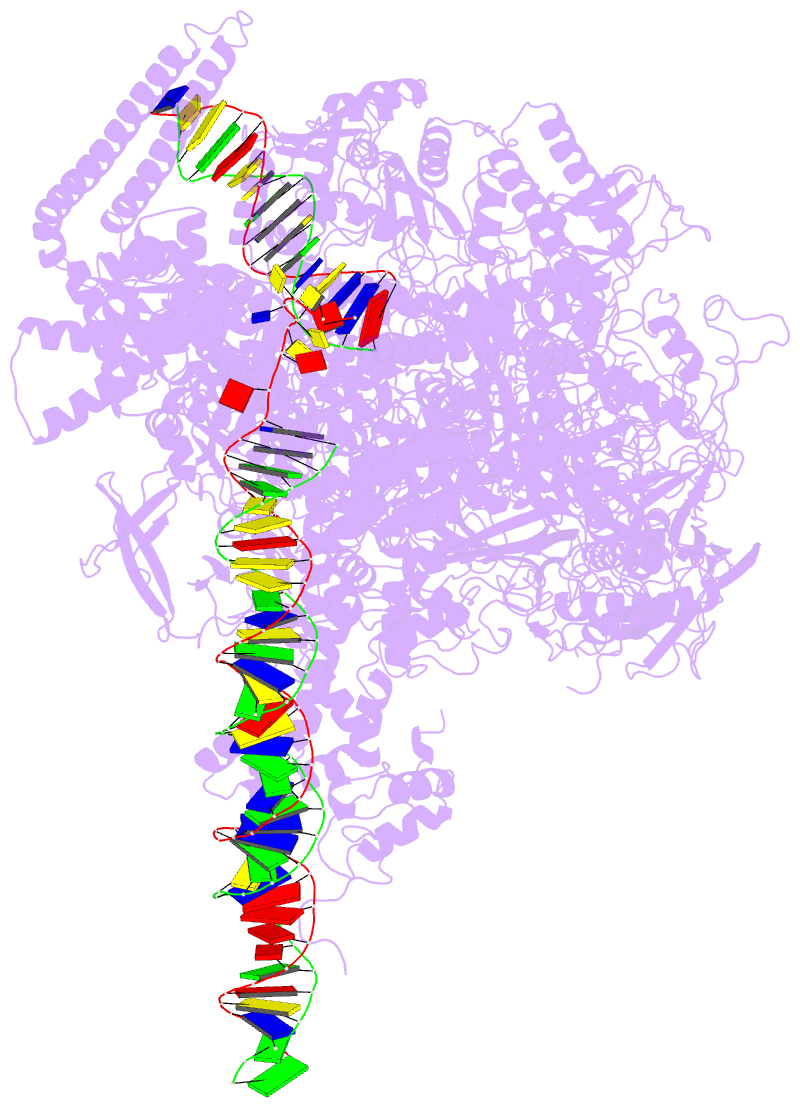
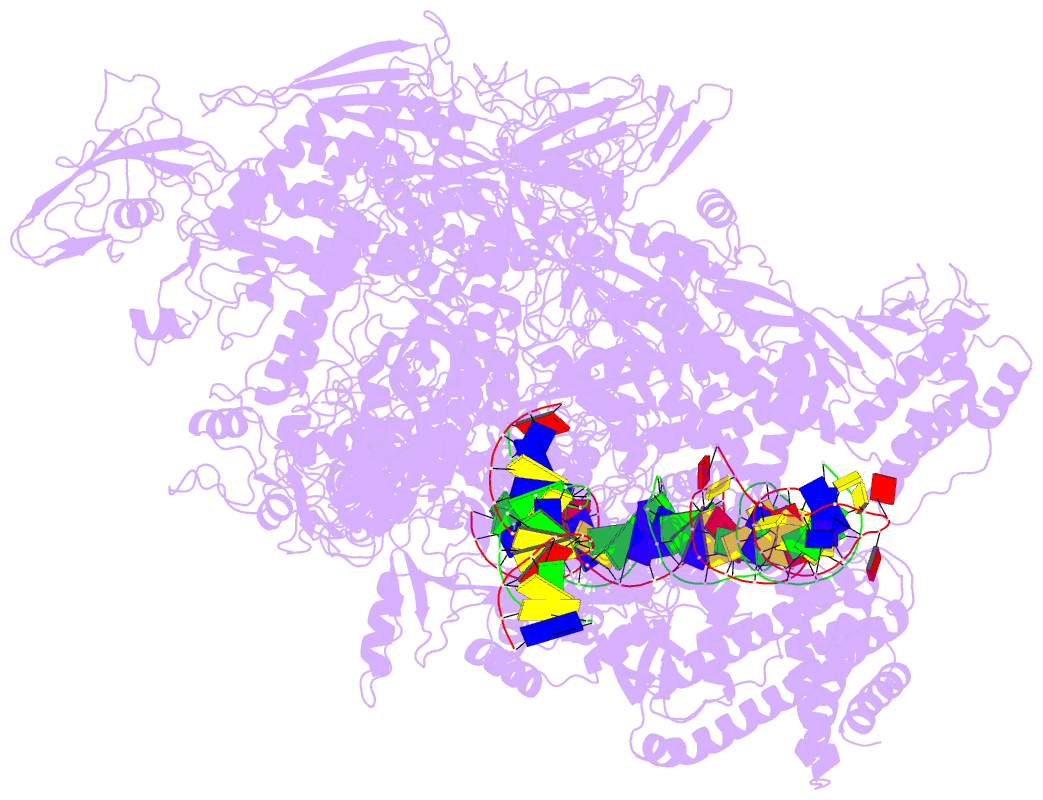
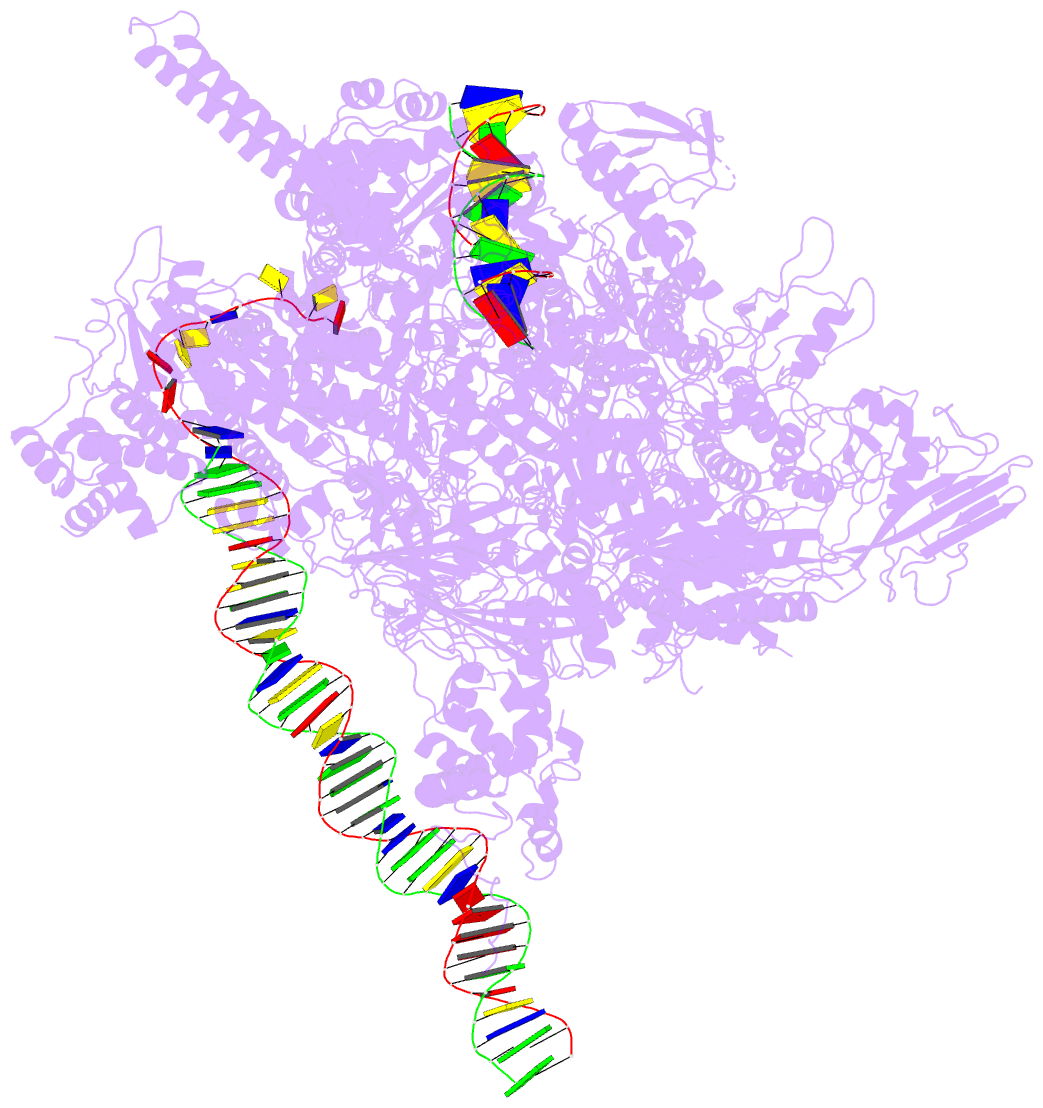
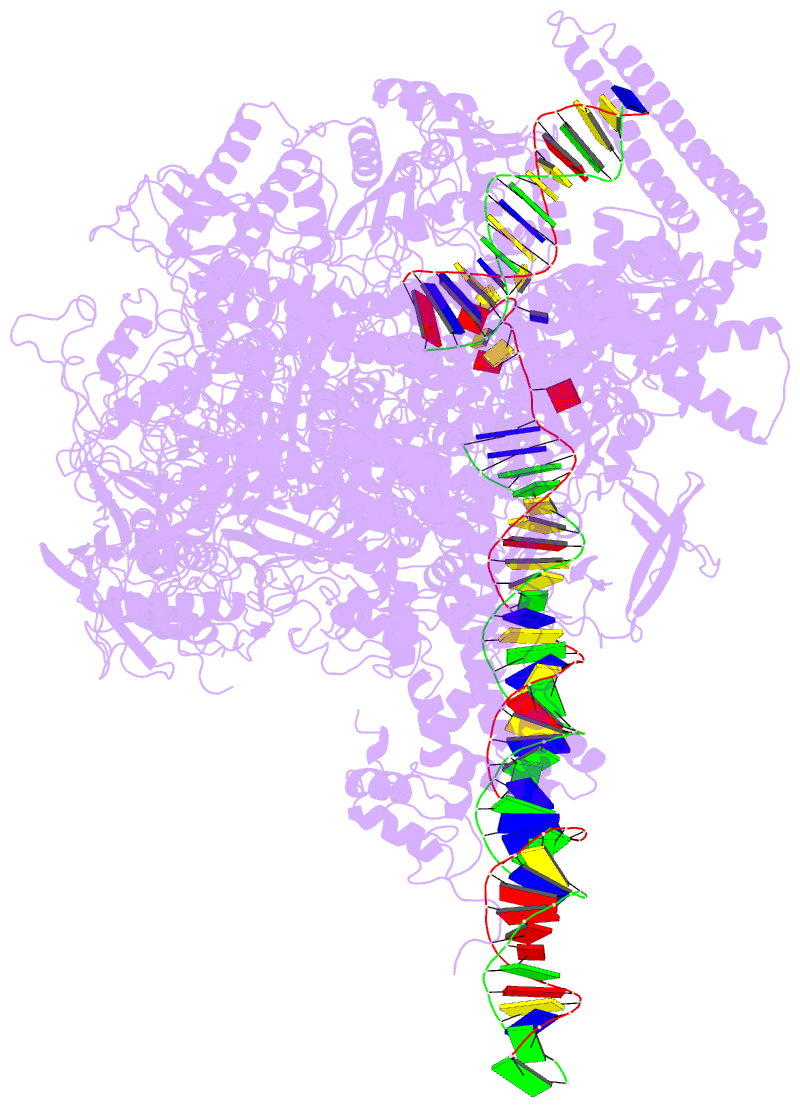
Summary information and primary citation
- PDB-id
-
7kif;
SNAP-derived features in text and
JSON formats
- Class
- transcription, transferase-DNA
- Method
- cryo-EM (2.94 Å)
- Summary
- Mycobacterium tuberculosis wt rnap transcription open
promoter complex with whib7 transcription factor
- Reference
-
Lilic M, Darst SA, Campbell EA (2021): "Structural
basis of transcriptional activation by the Mycobacterium
tuberculosis intrinsic antibiotic-resistance
transcription factor WhiB7." Mol.Cell,
81, 2875-2886.e5. doi: 10.1016/j.molcel.2021.05.017.
- Abstract
- In pathogenic mycobacteria, transcriptional responses
to antibiotics result in induced antibiotic resistance.
WhiB7 belongs to the Actinobacteria-specific family of
Fe-S-containing transcription factors and plays a crucial
role in inducible antibiotic resistance in mycobacteria.
Here, we present cryoelectron microscopy structures of
Mycobacterium tuberculosis transcriptional regulatory
complexes comprising RNA polymerase
σ<sub>A</sub>-holoenzyme, global regulators
CarD and RbpA, and WhiB7, bound to a WhiB7-regulated
promoter. The structures reveal how WhiB7 interacts with
σ<sub>A</sub>-holoenzyme while simultaneously
interacting with an AT-rich sequence element via its
AT-hook. Evidently, AT-hooks, rare elements in bacteria yet
prevalent in eukaryotes, bind to target AT-rich DNA
sequences similarly to the nuclear chromosome binding
proteins. Unexpectedly, a subset of particles contained a
WhiB7-stabilized closed promoter complex, revealing this
intermediate's structure, and we apply kinetic modeling and
biochemical assays to rationalize how WhiB7 activates
transcription. Altogether, our work presents a
comprehensive view of how WhiB7 serves to activate gene
expression leading to antibiotic resistance.