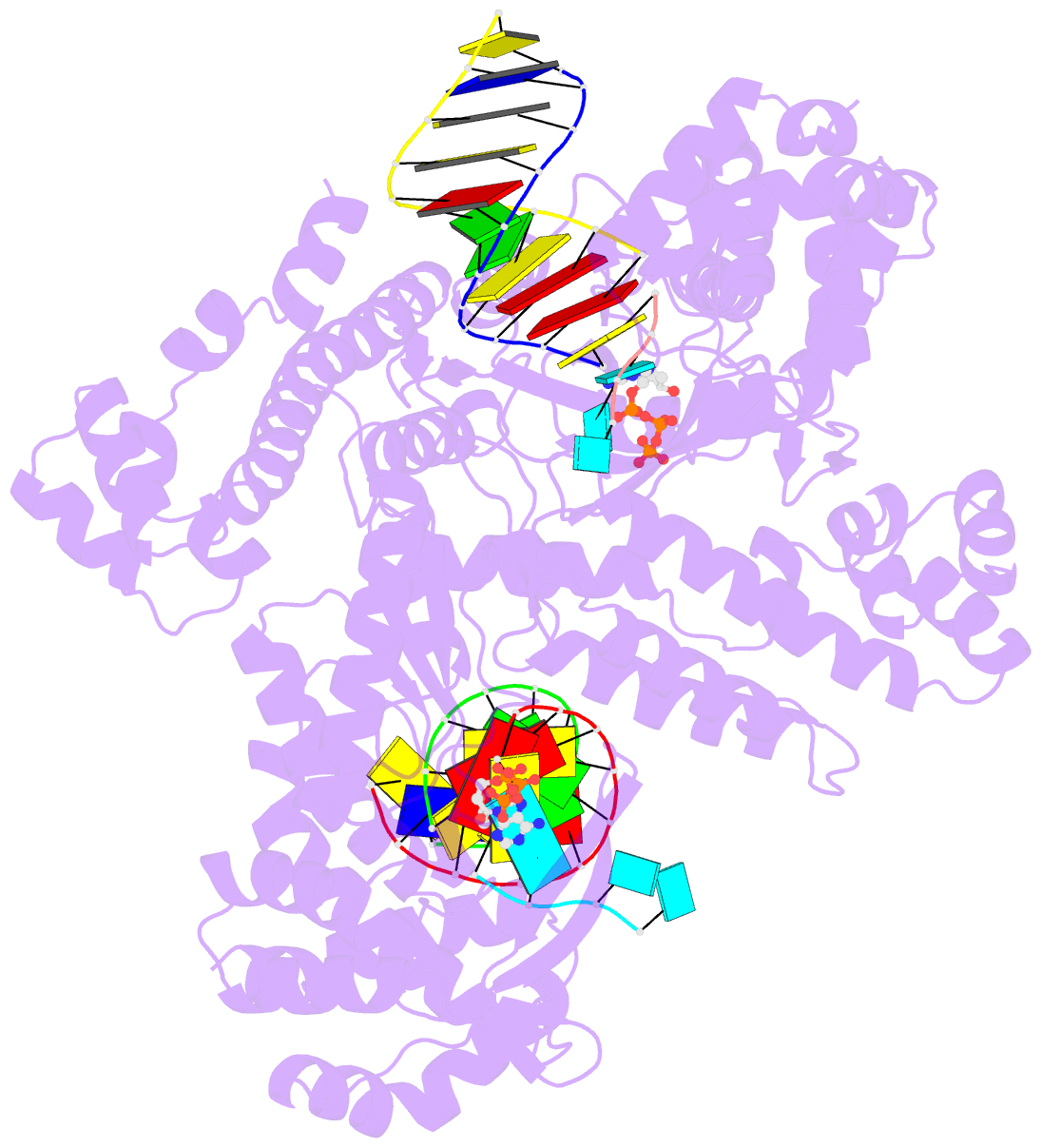
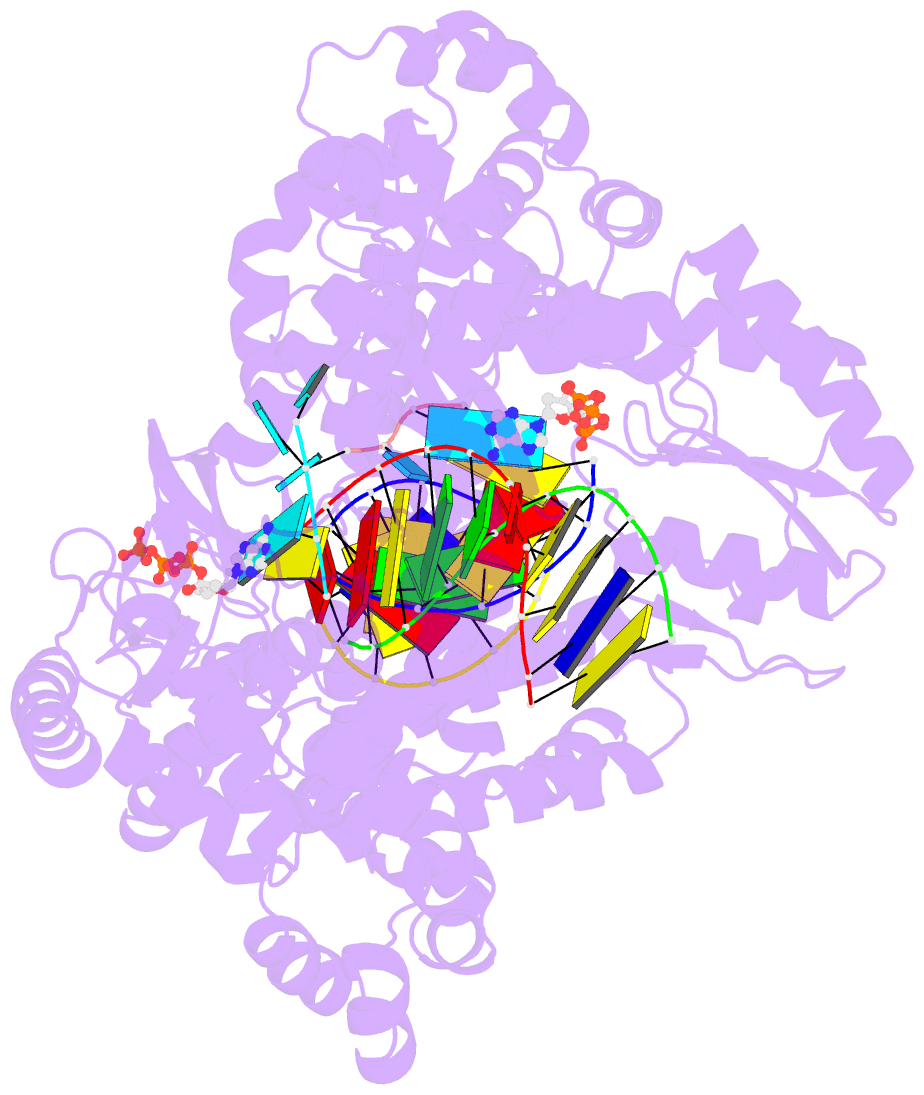
Summary information and primary citation
- PDB-id
- 7k9y; DSSR-derived features in text and JSON formats
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (3.2 Å)
- Summary
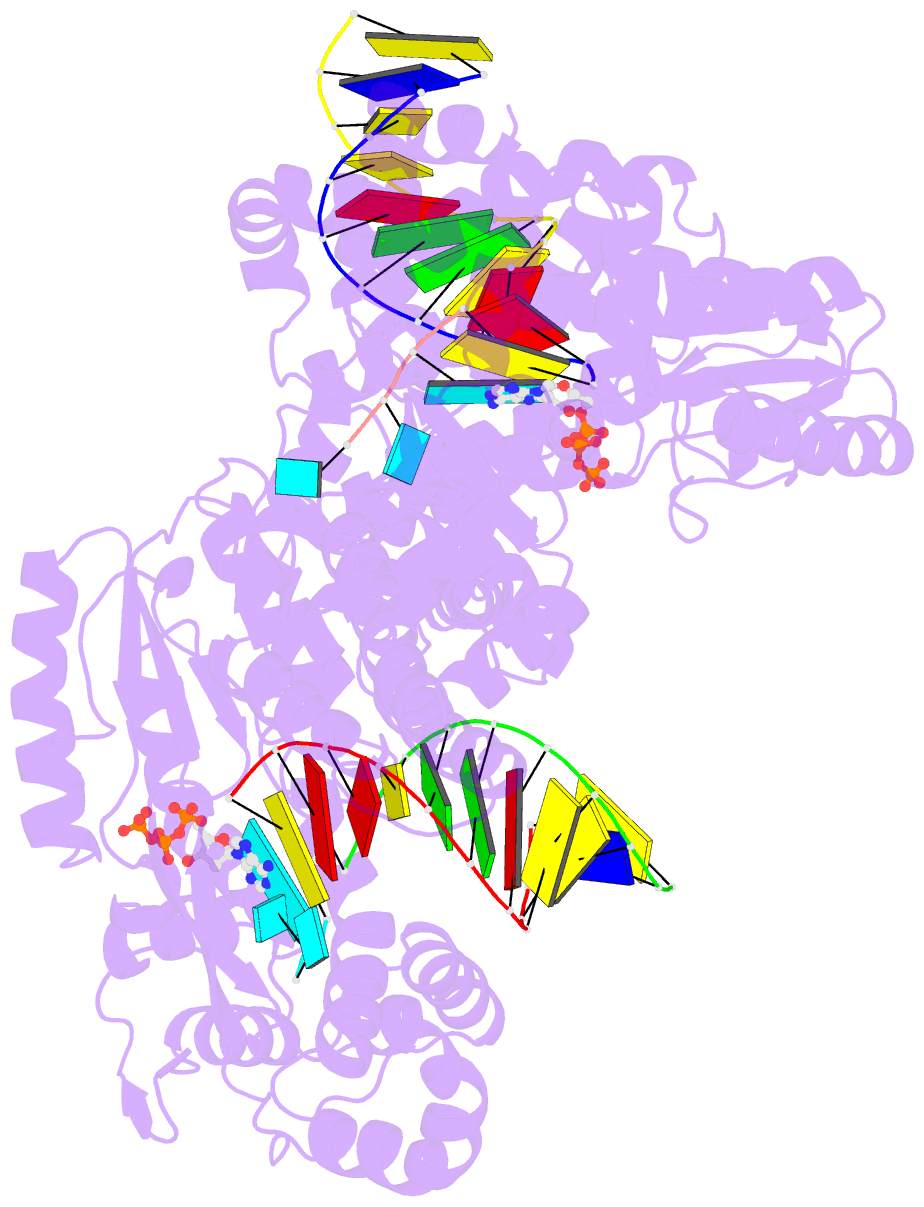
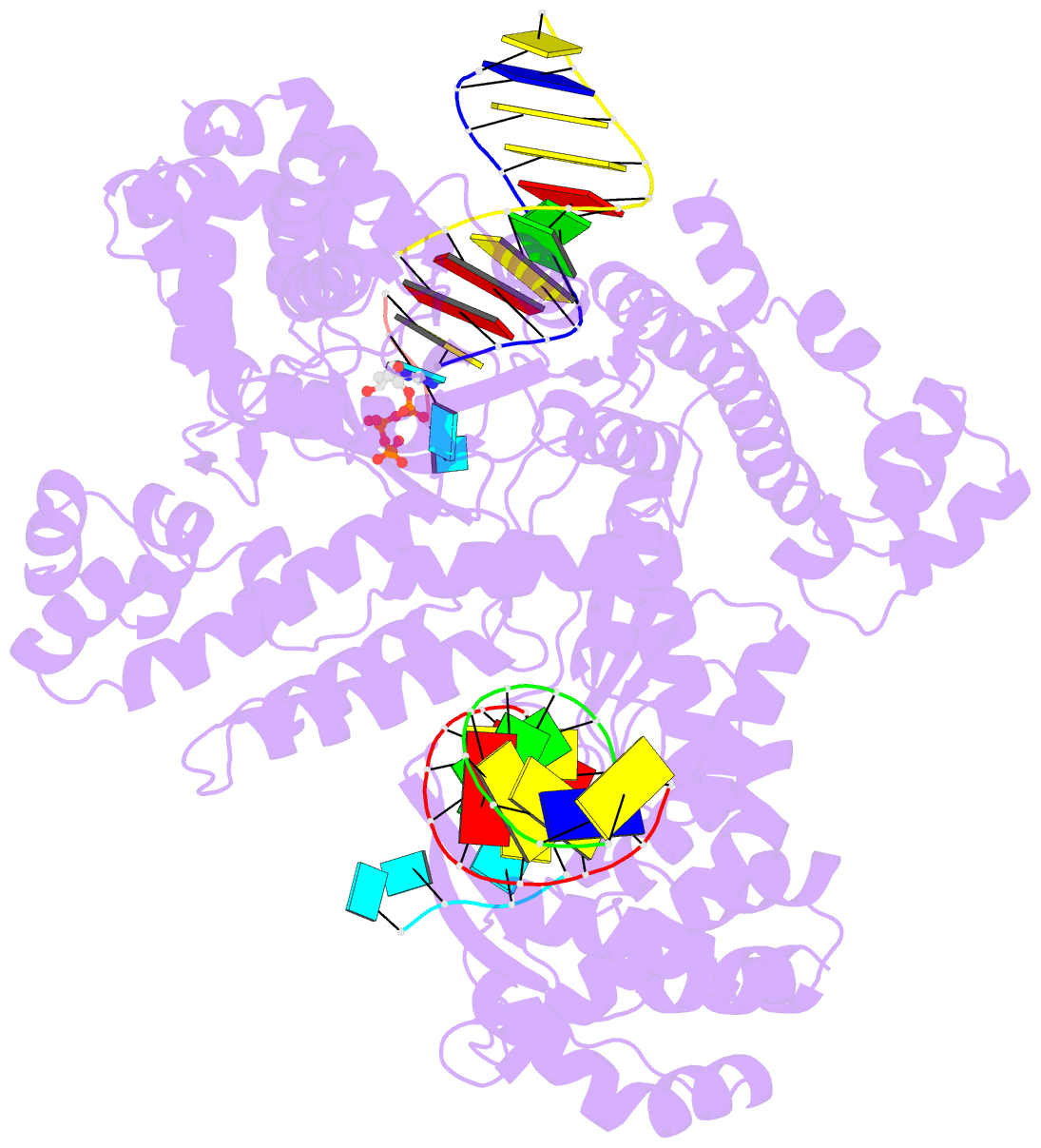
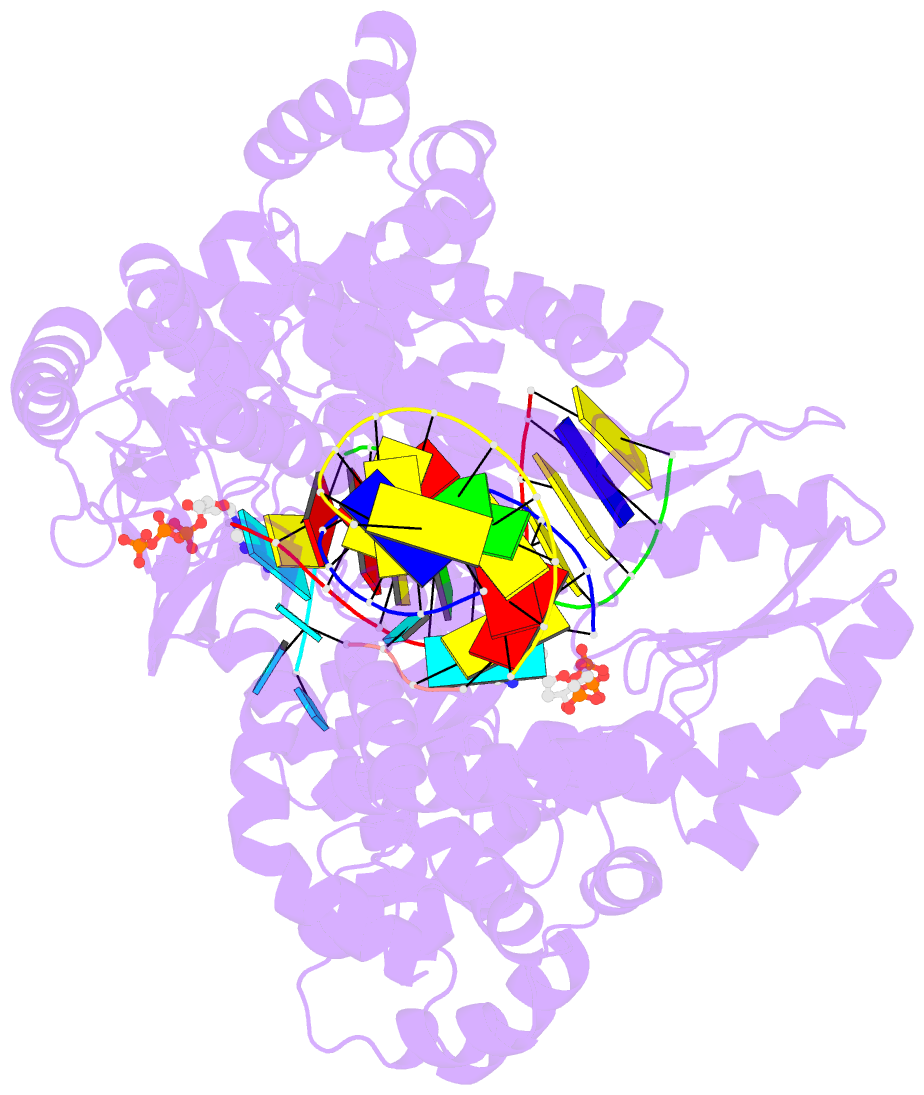
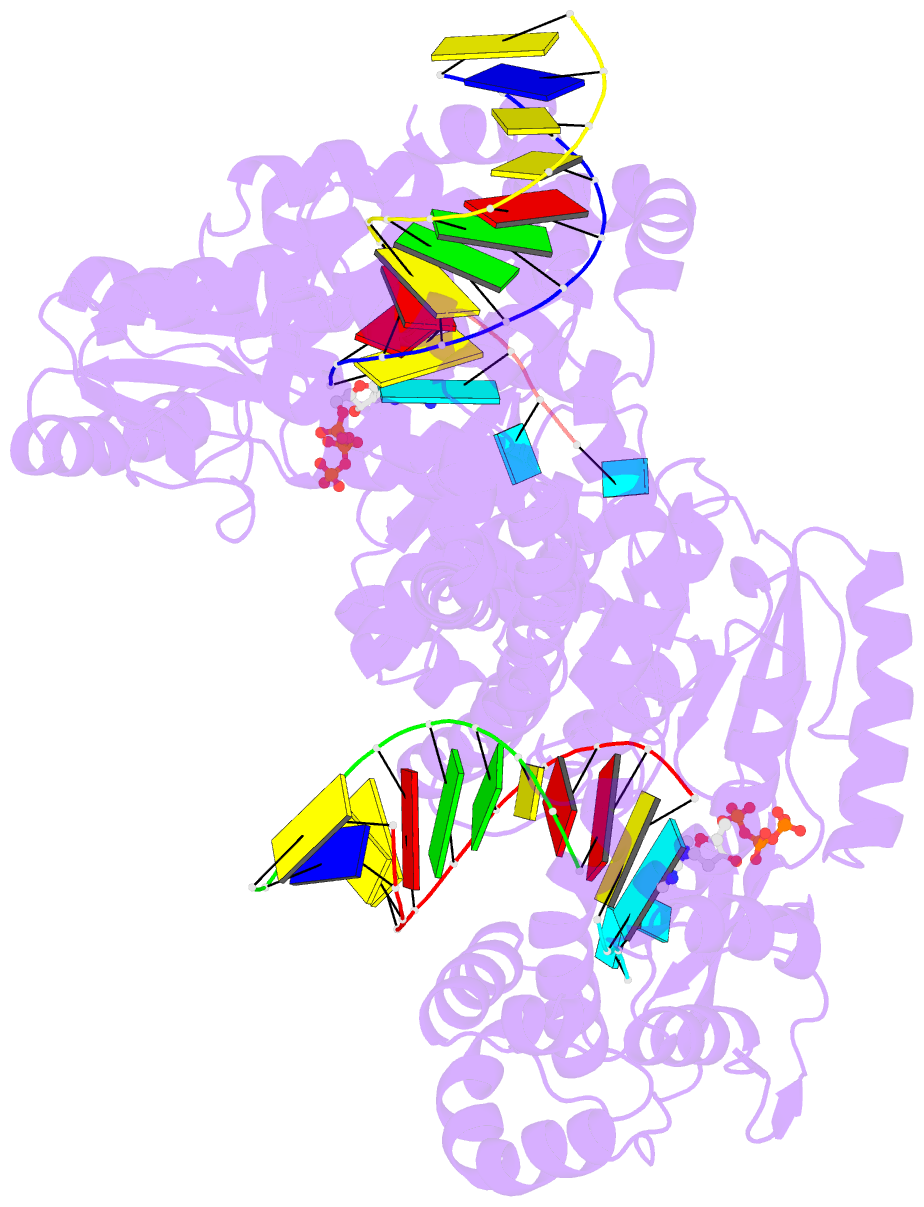
- Gsi-iic rt template-switching complex (twinned)
- Reference
- Lentzsch AM, Stamos JL, Yao J, Russell R, Lambowitz AM (2021): "Structural basis for template switching by a group II intron-encoded non-LTR-retroelement reverse transcriptase." J.Biol.Chem., 100971. doi: 10.1016/j.jbc.2021.100971.
- Abstract
- Reverse transcriptases (RTs) can switch template strands during cDNA synthesis, enabling them to join discontinuous nucleic acid sequences. Template switching plays crucial roles in retroviral replication and recombination, is used for adapter addition in RNA-seq, and may contribute to retroelement fitness by increasing evolutionary diversity and enabling continuous cDNA synthesis on damaged templates. Here, we determined an X-ray crystal structure of a template-switching complex of a group II intron RT bound simultaneously to an acceptor RNA and donor RNA template/DNA heteroduplex with a 1-nt 3'-DNA overhang. The structure showed that the 3' end of the acceptor RNA binds in a pocket formed by an N-terminal extension (NTE) present in non-long-terminal-repeat (LTR)-retroelement RTs and the RT fingertips loop, with the 3' nucleotide of the acceptor base paired to the 1-nt 3'-DNA overhang and its penultimate nucleotide base paired to the incoming dNTP at the RT active site. Analysis of structure-guided mutations identified amino acids that contribute to acceptor RNA binding and a phenylalanine residue near the RT active site that mediates non-templated addition (NTA). Mutation of the latter residue decreased multiple sequential template switches in RNA-seq. Our results provide new insights into the mechanisms of template switching and NTA by RTs, suggest how these reactions could be improved for RNA-seq, and reveal common structural features for template switching by non-LTR-retroelement RTs and viral RNA-dependent RNA polymerases.