Summary information and primary citation
- PDB-id
-
7e1b;
SNAP-derived features in text and
JSON formats
- Class
- DNA binding protein
- Method
- X-ray (4.587 Å)
- Summary
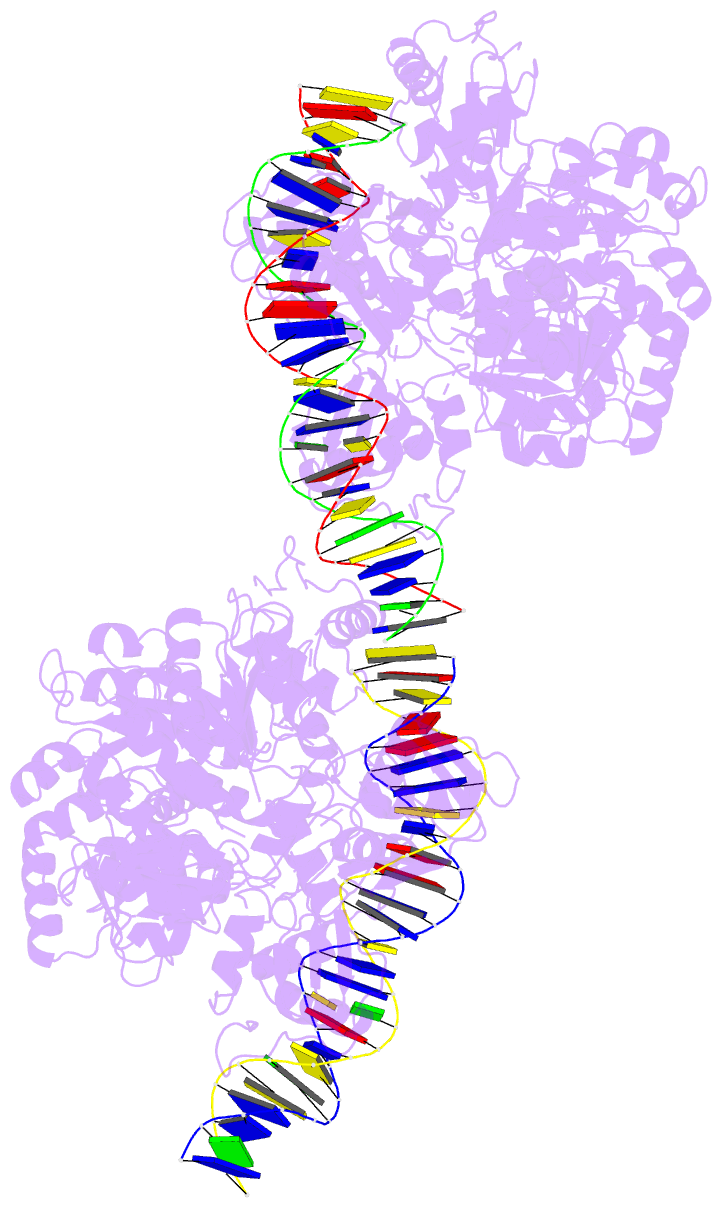
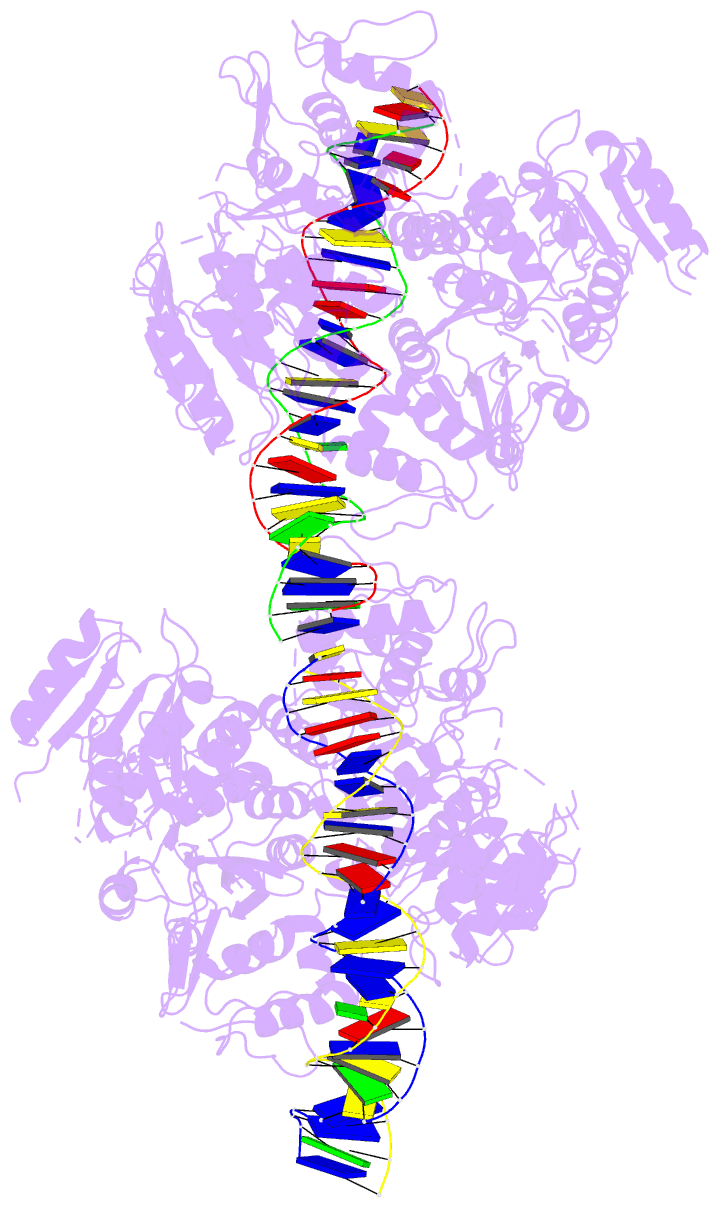
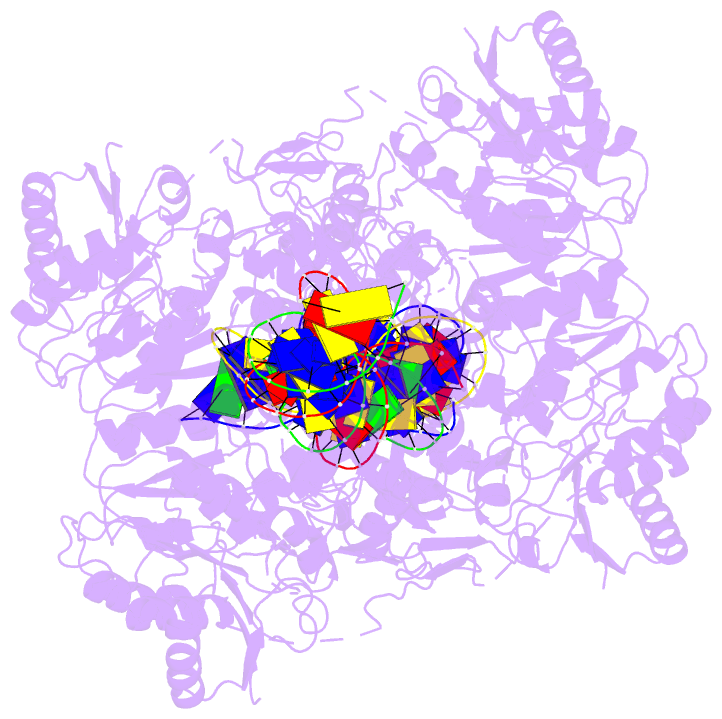
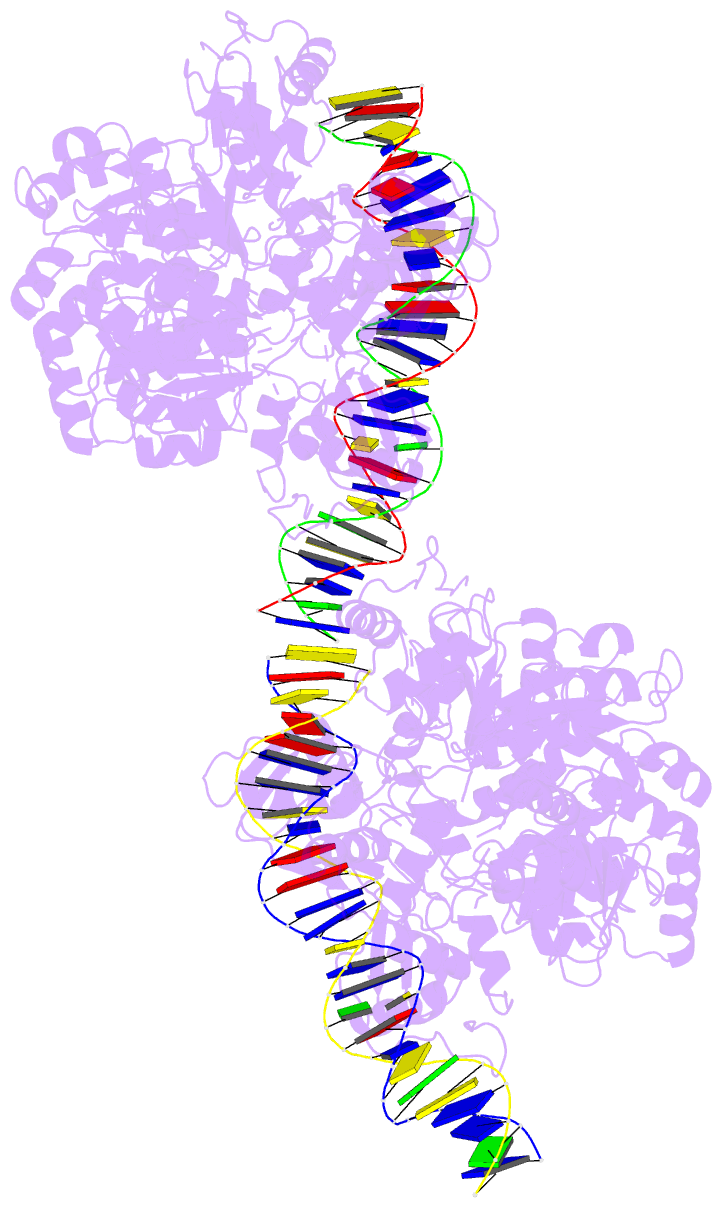
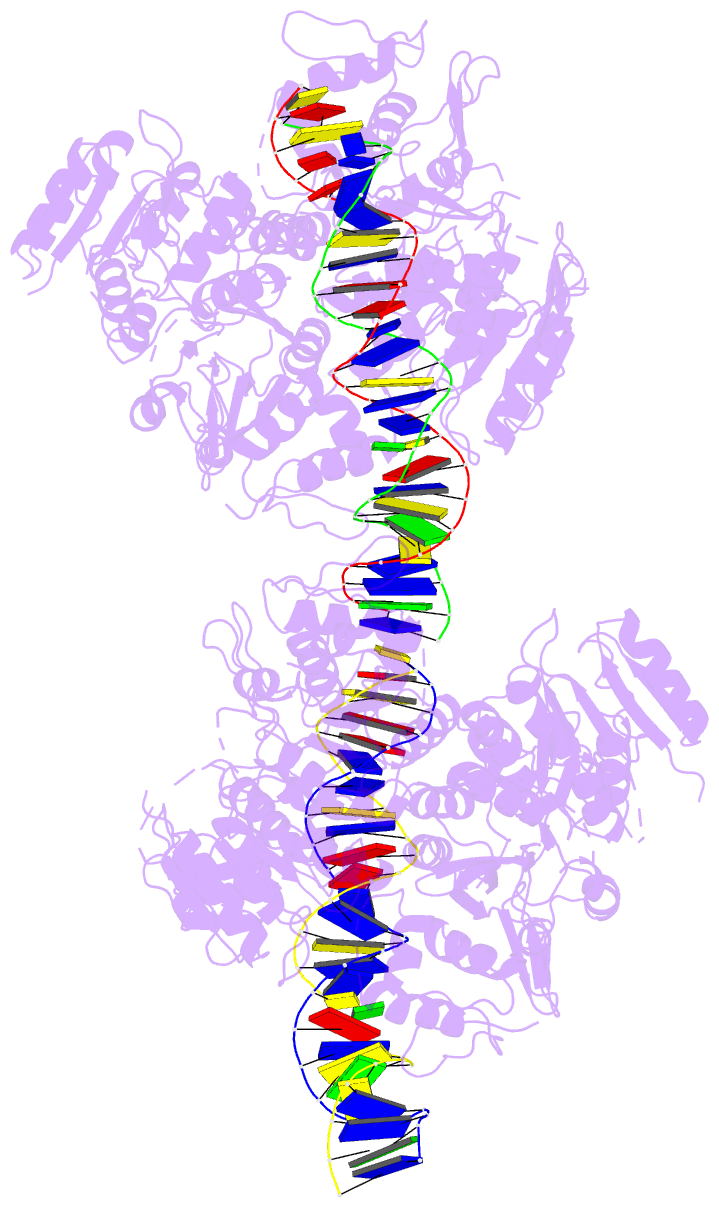
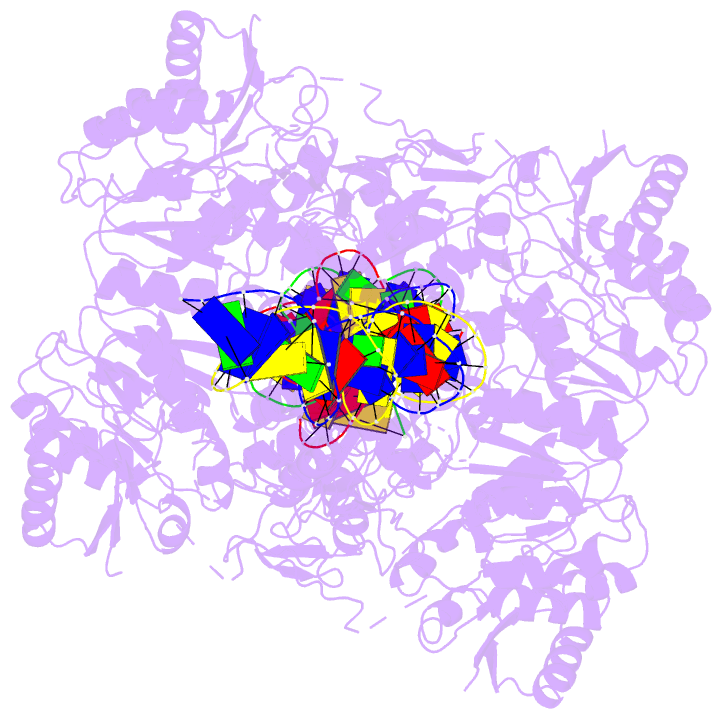
- Crystal structure of vbrr-DNA complex
- Reference
-
Hong S, Guo J, Zhang X, Zhou X, Zhang P, Yu F (2023):
"Structural
basis of phosphorylation-induced activation of the
response regulator VbrR." Acta
Biochim.Biophys.Sin. doi: 10.3724/abbs.2022200.
- Abstract
- <p indent="0mm">Two-component systems typically
consist of a paired histidine kinase and response regulator
and couple environmental changes to adaptive responses. The
response regulator VbrR from <italic>Vibrio
parahaemolyticus</italic>, a member of the OmpR/PhoB
family, regulates virulence and antibiotic resistance
genes. The activation mechanism of VbrR remains unclear.
Here, we report the crystal structures of full-length VbrR
in complex with DNA in the active conformation and the
N-terminal receiver domain (RD) and the C-terminal
DNA-binding domain (DBD) in both active and inactive
conformations. Structural and biochemical analyses suggest
that unphosphorylated VbrR adopts mainly as inactive dimers
through the DBD at the autoinhibitory state. The RD
undergoes a monomer-to-dimer transition upon
phosphorylation, which further induces the transition of
DBD from an autoinhibitory dimer to an active dimer and
enables its binding with target DNA. Our study suggests a
new model for phosphorylation-induced activation of
response regulators and sheds light on the pathogenesis of
<italic>V</italic>.
<italic>parahaemolyticus</italic>.
</p>.