Summary information and primary citation
- PDB-id
- 7b0y; DSSR-derived features in text and JSON formats
- Class
- splicing
- Method
- cryo-EM (3.6 Å)
- Summary
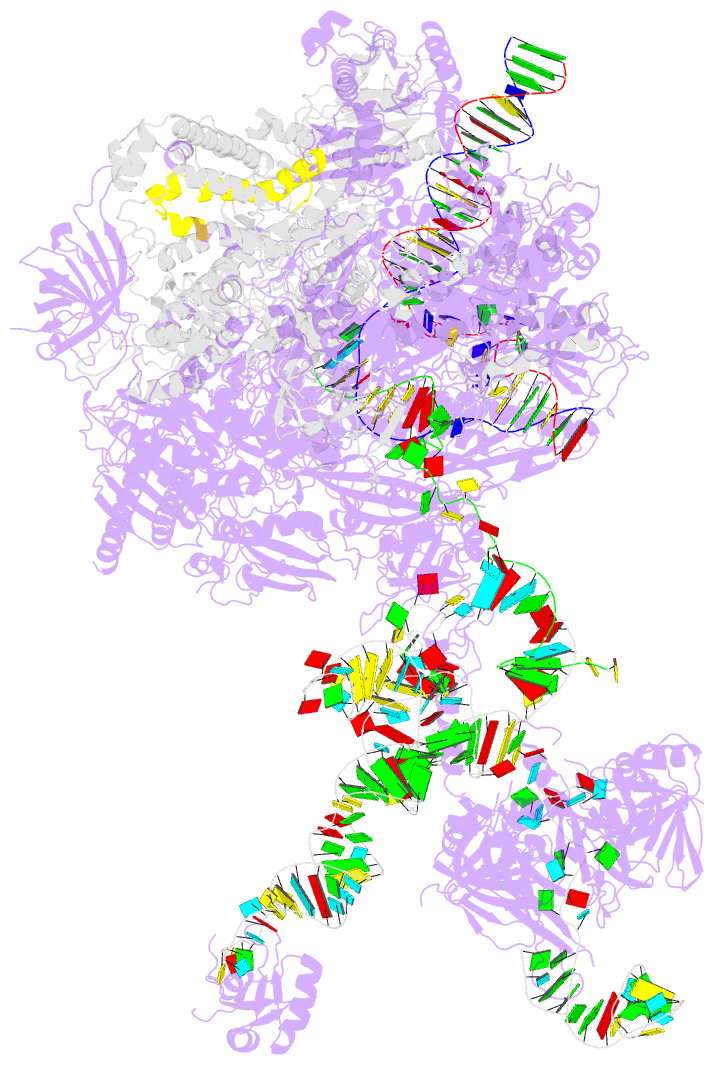
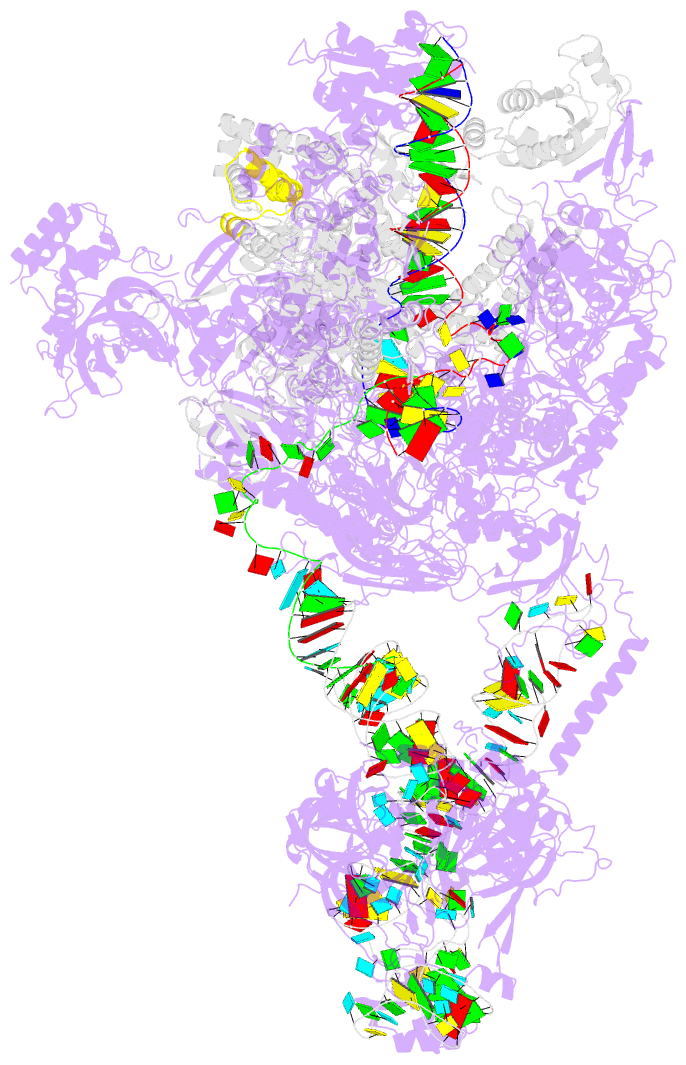
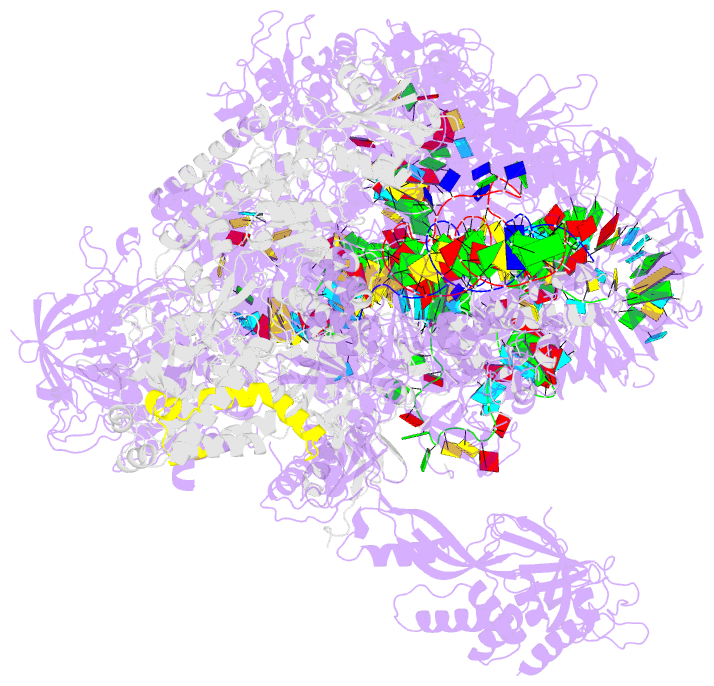
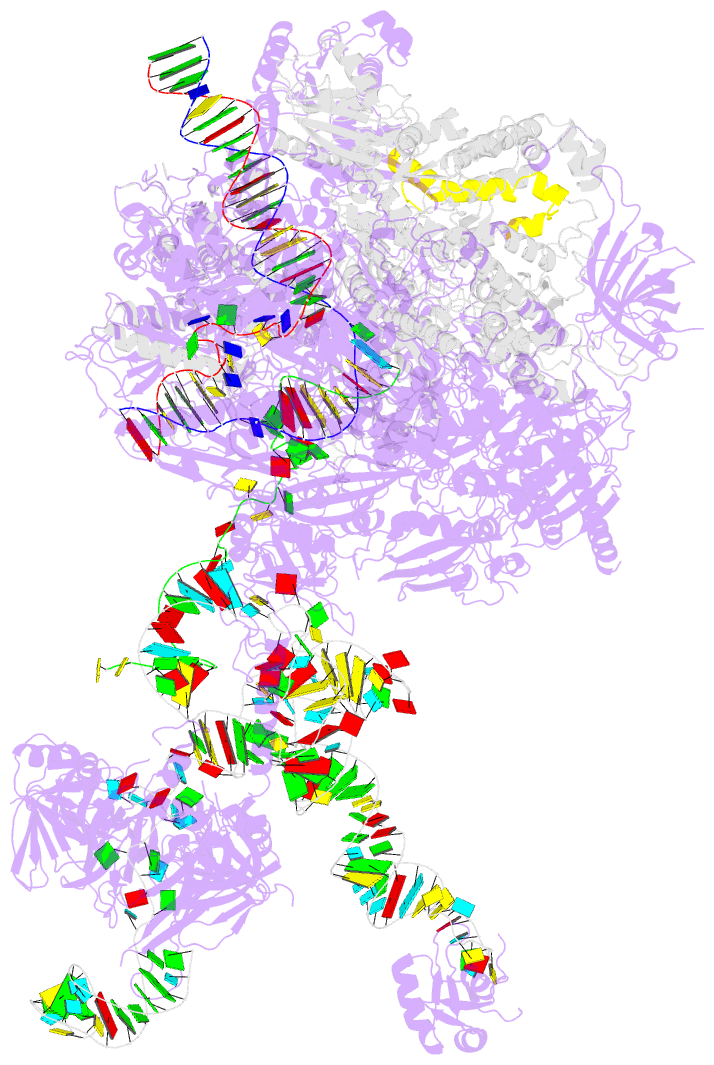
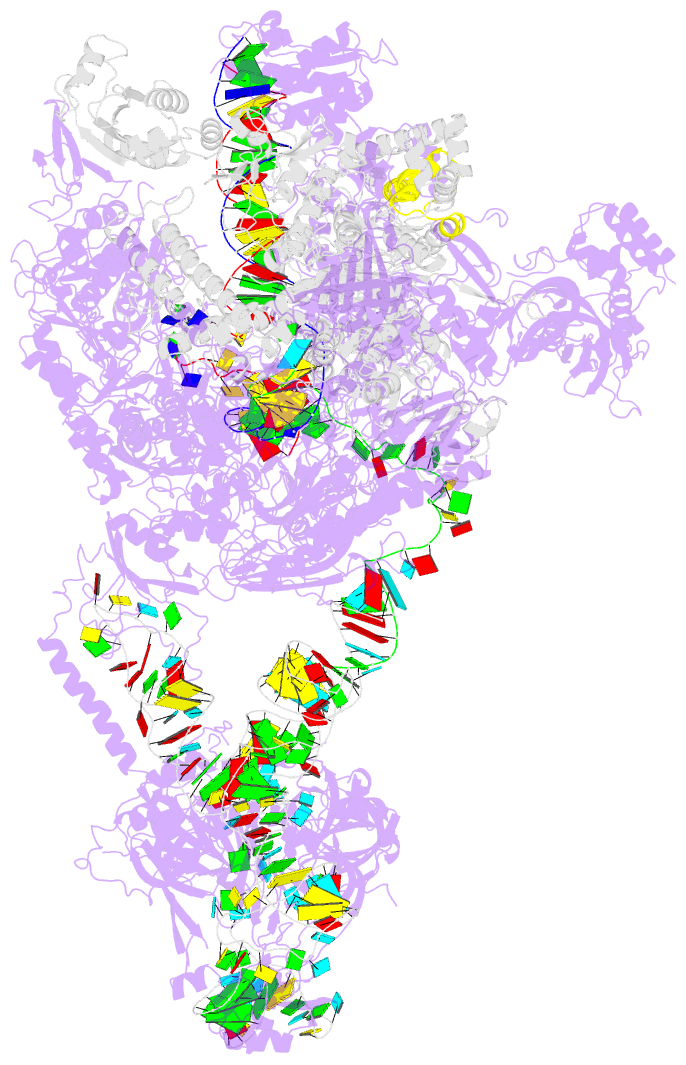
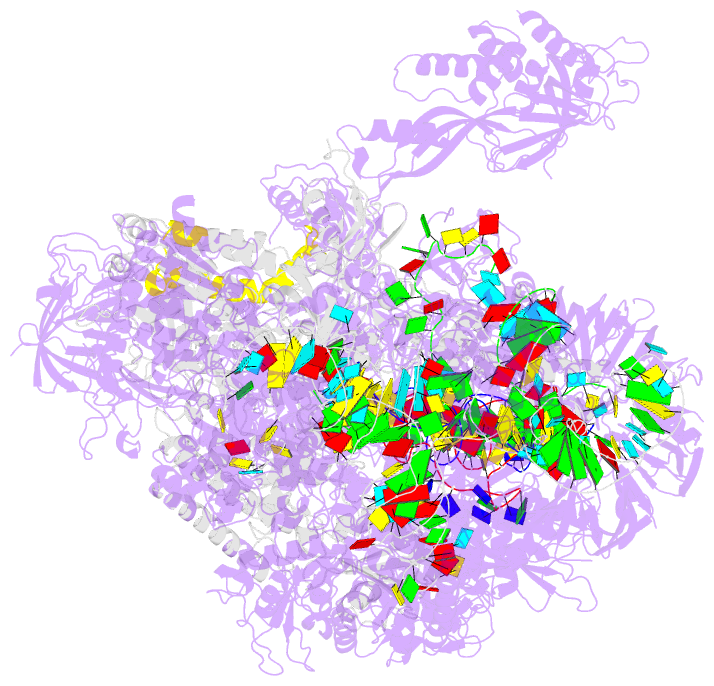
- Structure of a transcribing RNA polymerase ii-u1 snrnp complex
- Reference
- Zhang S, Aibara S, Vos SM, Agafonov DE, Luhrmann R, Cramer P (2021): "Structure of a transcribing RNA polymerase II-U1 snRNP complex." Science, 371, 305-309. doi: 10.1126/science.abf1870.
- Abstract
- To initiate cotranscriptional splicing, RNA polymerase II (Pol II) recruits the U1 small nuclear ribonucleoprotein particle (U1 snRNP) to nascent precursor messenger RNA (pre-mRNA). Here, we report the cryo-electron microscopy structure of a mammalian transcribing Pol II-U1 snRNP complex. The structure reveals that Pol II and U1 snRNP interact directly. This interaction positions the pre-mRNA 5' splice site near the RNA exit site of Pol II. Extension of pre-mRNA retains the 5' splice site, leading to the formation of a "growing intron loop." Loop formation may facilitate scanning of nascent pre-mRNA for the 3' splice site, functional pairing of distant intron ends, and prespliceosome assembly. Our results provide a starting point for a mechanistic analysis of cotranscriptional spliceosome assembly and the biogenesis of mRNA isoforms by alternative splicing.