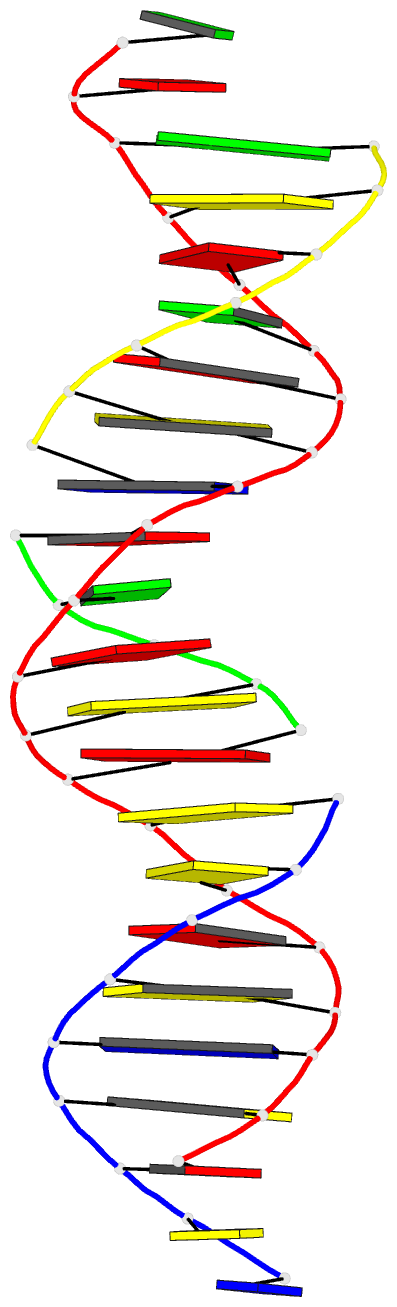
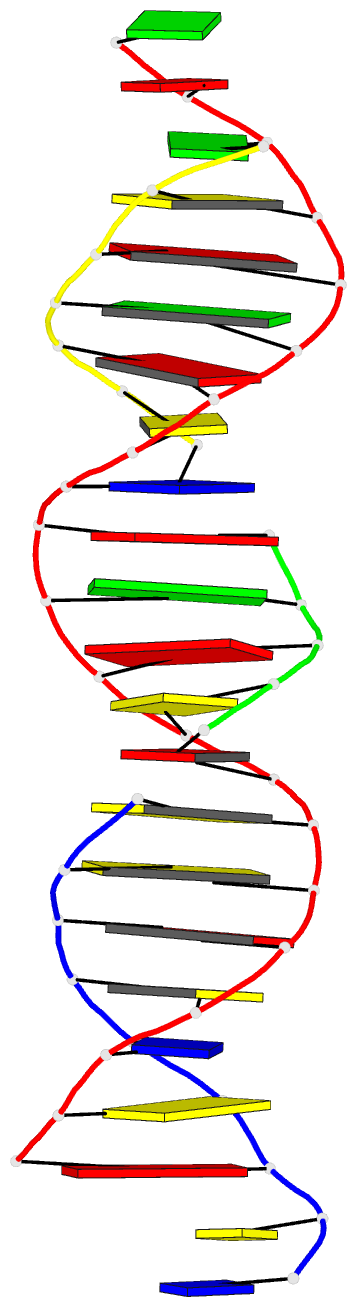
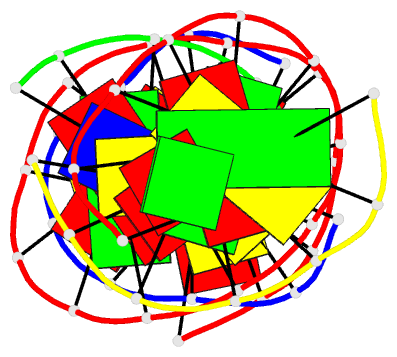
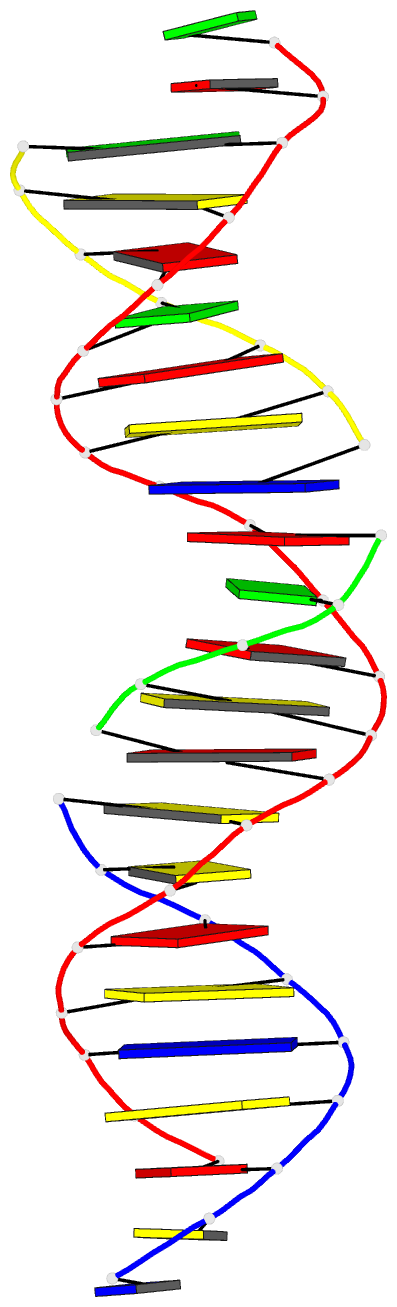
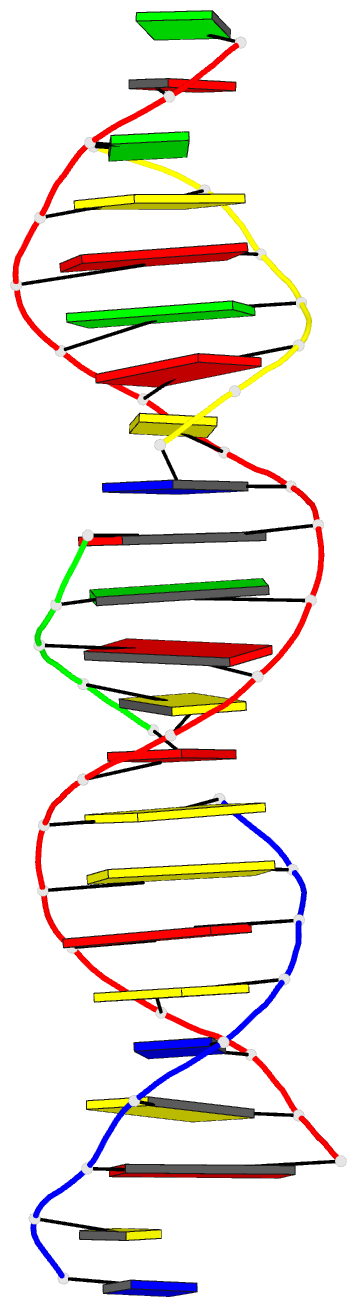
Summary information and primary citation
- PDB-id
- 6wr9; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (3.071 Å)
- Summary
- Self-assembly of a 3d DNA crystal lattice (4x5 duplex version) containing the j32 immobile holliday junction
- Reference
- Simmons CR, MacCulloch T, Krepl M, Matthies M, Buchberger A, Crawford I, Sponer J, Sulc P, Stephanopoulos N, Yan H (2022): "The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly." Nat Commun, 13, 3112. doi: 10.1038/s41467-022-30779-6.
- Abstract
- The programmable synthesis of rationally engineered crystal architectures for the precise arrangement of molecular species is a foundational goal in nanotechnology, and DNA has become one of the most prominent molecules for the construction of these materials. In particular, branched DNA junctions have been used as the central building block for the assembly of 3D lattices. Here, crystallography is used to probe the effect of all 36 immobile Holliday junction sequences on self-assembling DNA crystals. Contrary to the established paradigm in the field, most junctions yield crystals, with some enhancing the resolution or resulting in unique crystal symmetries. Unexpectedly, even the sequence adjacent to the junction has a significant effect on the crystal assemblies. Six of the immobile junction sequences are completely resistant to crystallization and thus deemed "fatal," and molecular dynamics simulations reveal that these junctions invariably lack two discrete ion binding sites that are pivotal for crystal formation. The structures and dynamics detailed here could be used to inform future designs of both crystals and DNA nanostructures more broadly, and have potential implications for the molecular engineering of applied nanoelectronics, nanophotonics, and catalysis within the crystalline context.