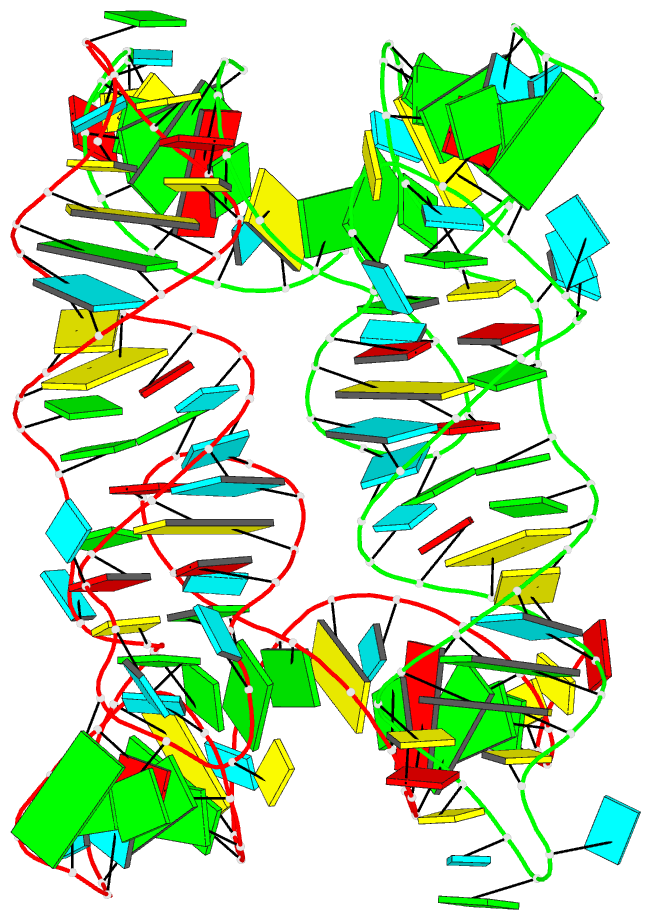
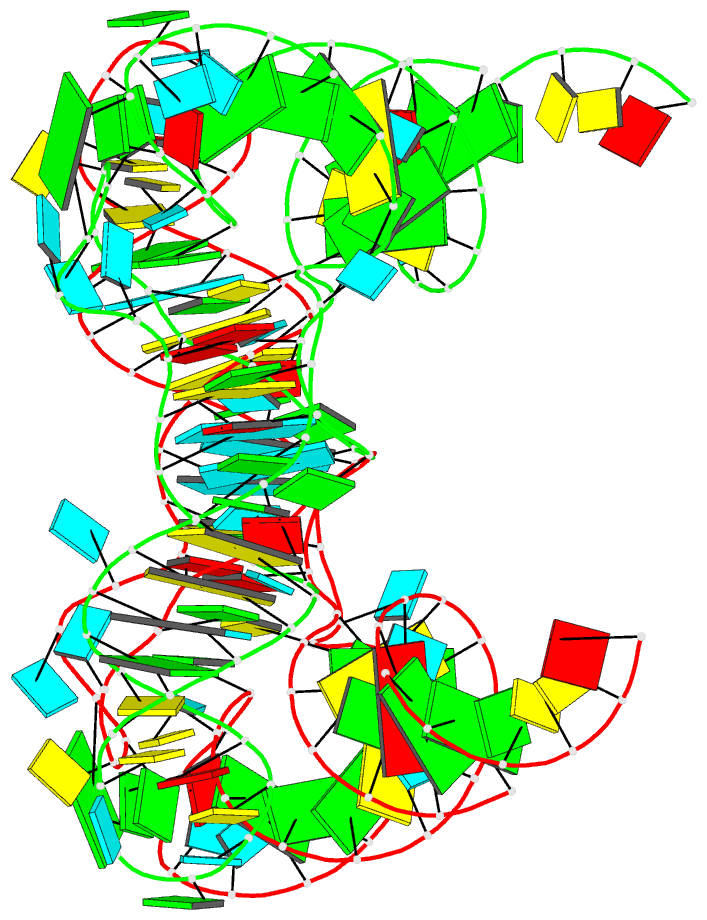
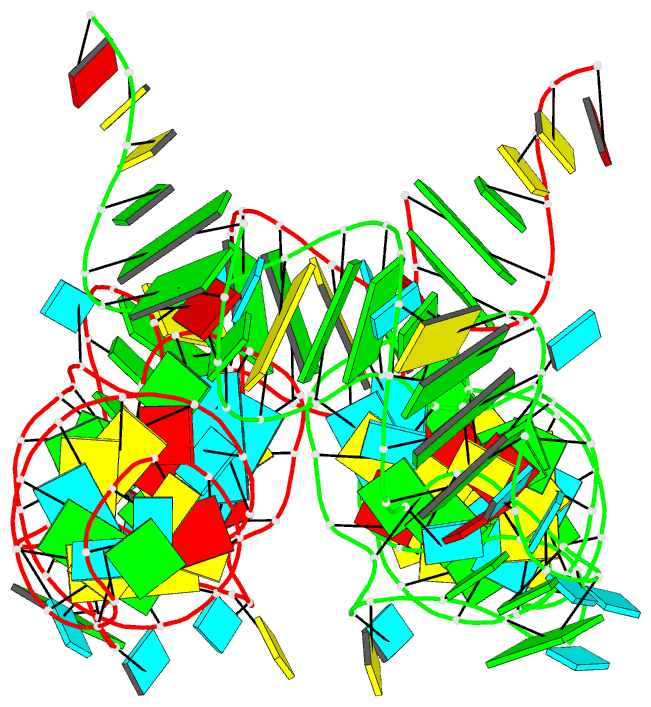
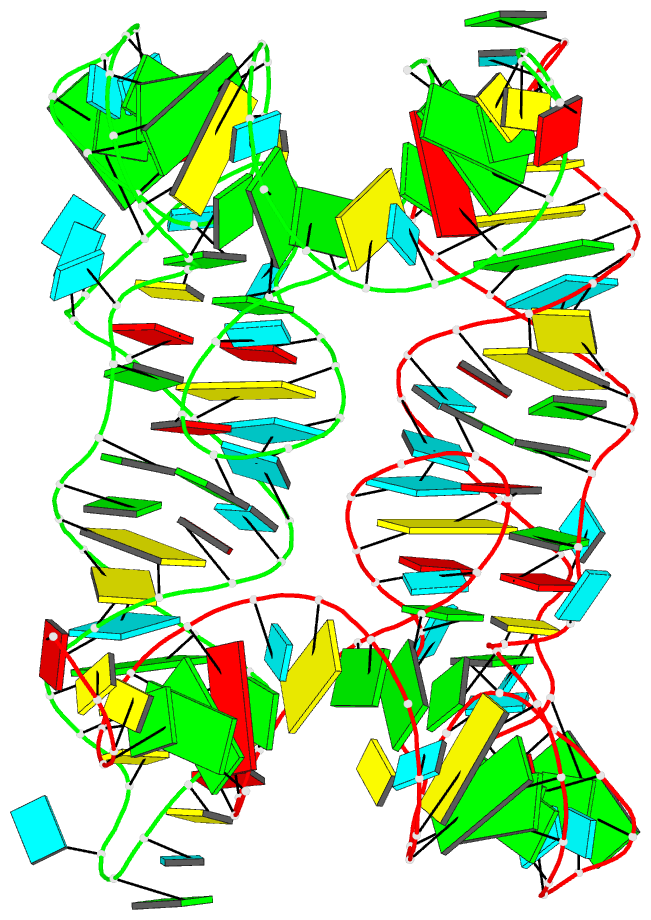
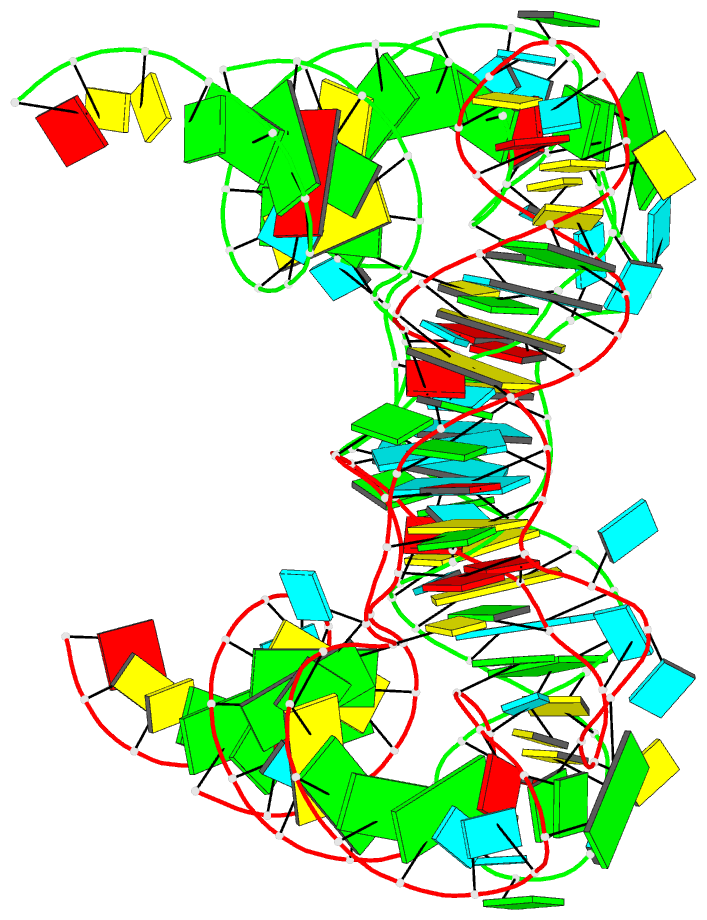
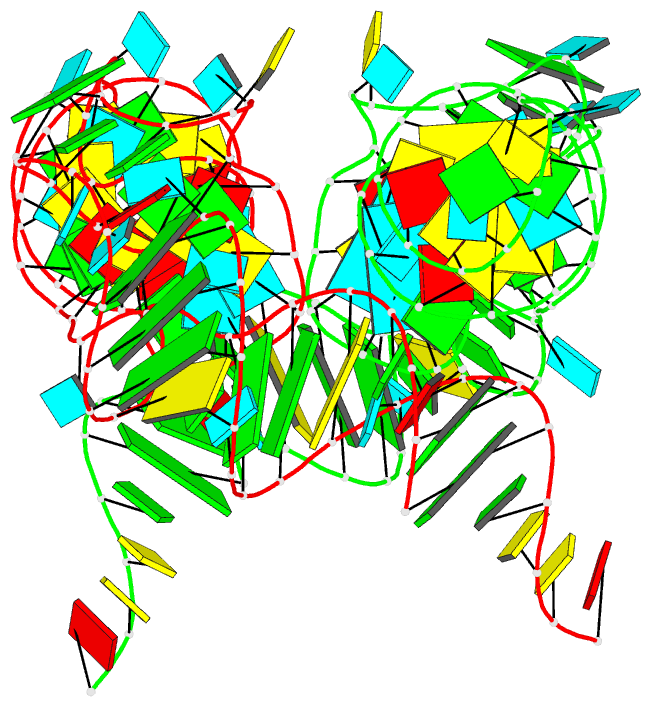
Summary information and primary citation
- PDB-id
- 6ugg; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (1.95 Å)
- Summary
- Structure of unmodified e. coli trna(asp)
- Reference
- Chan CW, Badong D, Rajan R, Mondragon A (2020): "Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs." Rna, 26, 278-289. doi: 10.1261/rna.073478.119.
- Abstract
- Ubiquitous across all domains of life, tRNAs constitute an essential component of cellular physiology, carry out an indispensable role in protein synthesis, and have been historically the subject of a wide range of biochemical and biophysical studies as prototypical folded RNA molecules. Although conformational flexibility is a well-established characteristic of tRNA structure, it is typically regarded as an adaptive property exhibited in response to an inducing event, such as the binding of a tRNA synthetase or the accommodation of an aminoacyl-tRNA into the ribosome. In this study, we present crystallographic data of a tRNA molecule to expand on this paradigm by showing that structural flexibility and plasticity are intrinsic properties of tRNAs, apparent even in the absence of other factors. Based on two closely related conformations observed within the same crystal, we posit that unbound tRNAs by themselves are flexible and dynamic molecules. Furthermore, we demonstrate that the formation of the T-loop conformation by the tRNA TΨC stem-loop, a well-characterized and classic RNA structural motif, is possible even in the absence of important interactions observed in fully folded tRNAs.