Summary information and primary citation
- PDB-id
-
6tb7;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (2.53 Å)
- Summary
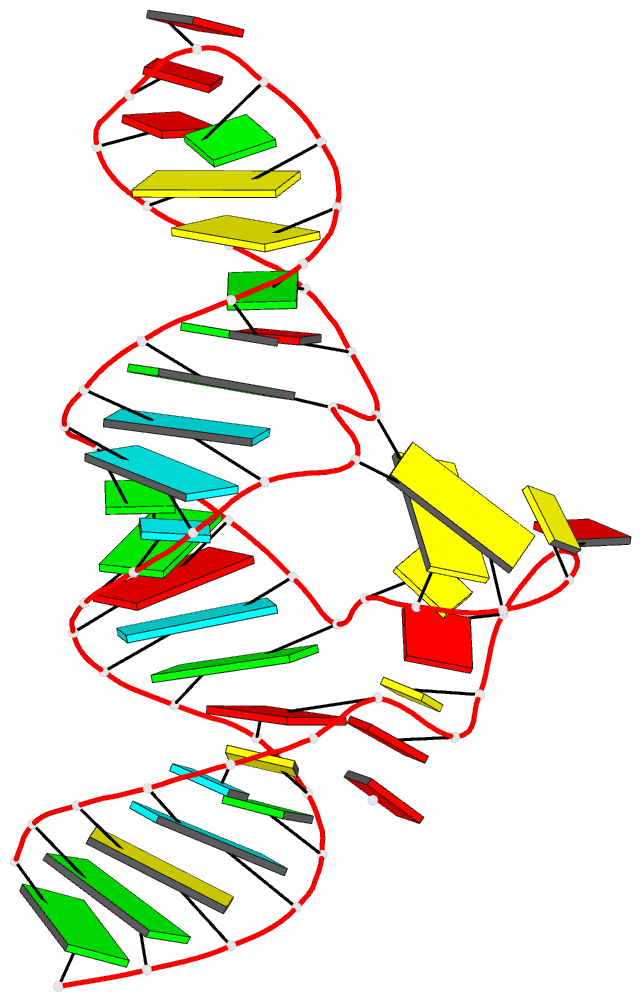
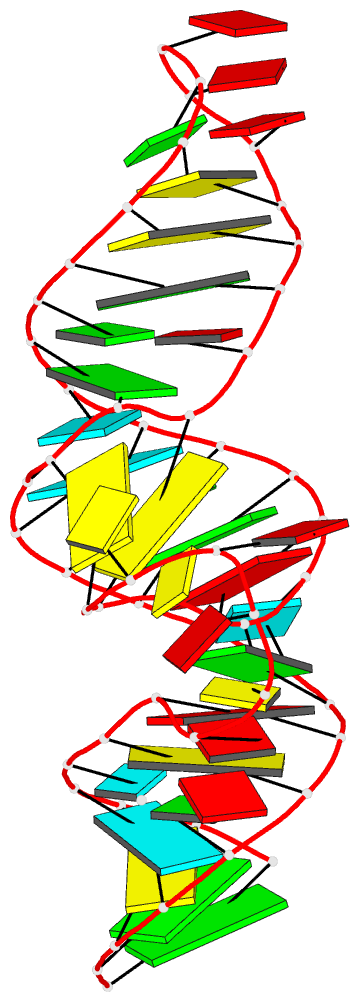
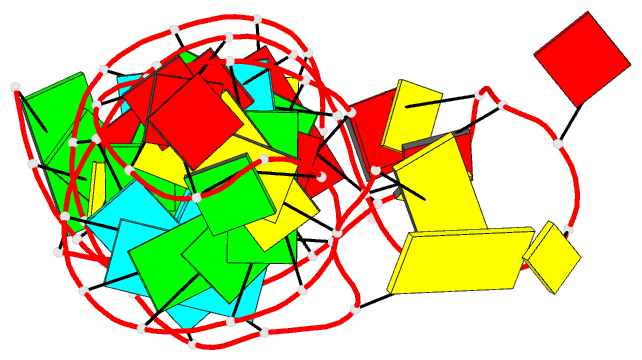
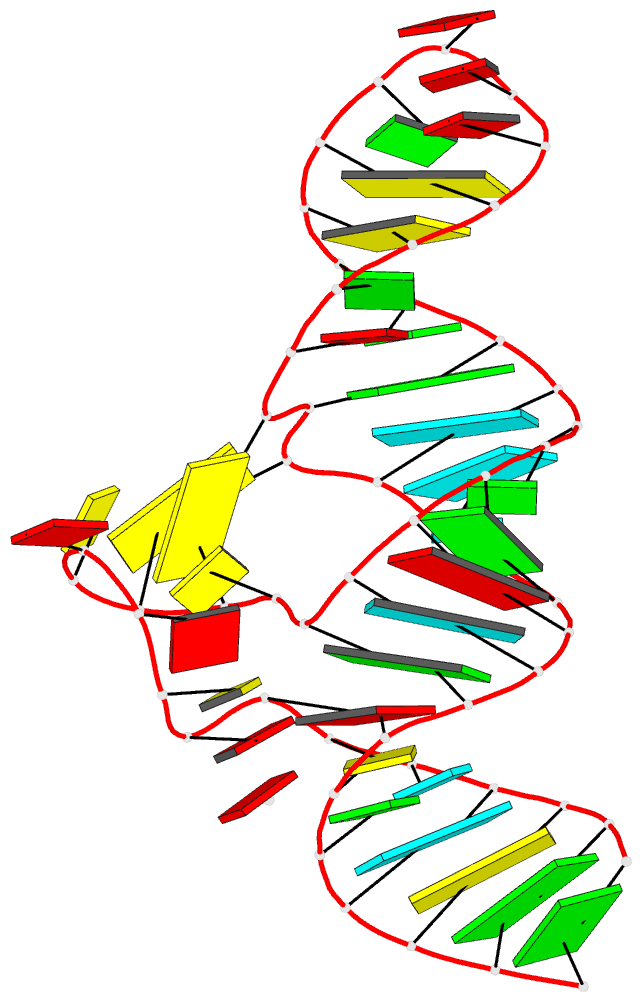
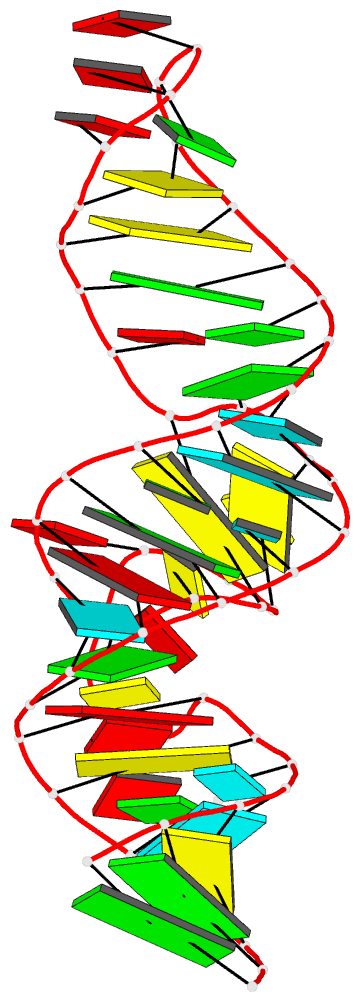
- Crystal structure of the adp-binding domain of the nad+
riboswitch with adenosine monophosphate (amp)
- Reference
-
Huang L, Wang J, Lilley DMJ (2020): "Structure
and ligand binding of the ADP-binding domain of the NAD +
riboswitch." Rna, 26,
878-887. doi: 10.1261/rna.074898.120.
- Abstract
- The <i>nad</i>A motif is the first known
NAD<sub>+</sub>-dependent riboswitch,
comprising two similar tandem bulged stem-loop structures.
We have determined the structure of the 5' domain 1 of the
riboswitch. It has three coaxial helical segments,
separated by an ACANCCCC bulge and by an internal loop,
with a tertiary contact between them that includes two C:G
base pairs. We have determined the structure with a number
of ligands related to NADH, but in each case only the ADP
moiety is observed. The adenosine adopts an
<i>anti</i> conformation, forms multiple
hydrogen bonds across the width of the sugar edge of the
penultimate C:G base pair of the helix preceding the bulge,
and the observed contacts have been confirmed by
mutagenesis and calorimetry. Two divalent metal ions play a
key structural role at the narrow neck of the bulge. One
makes direct bonding contacts to the diphosphate moiety,
locking it into position. Thus the nucleobase, ribose, and
phosphate groups of the ADP moiety are all specifically
recognized by the RNA. The NAD<sub>+</sub>
riboswitch is modular. Domain 1 is an ADP binding domain
that may be ancient and could potentially be used in
combination with other ligand binding motifs such as
CoA.