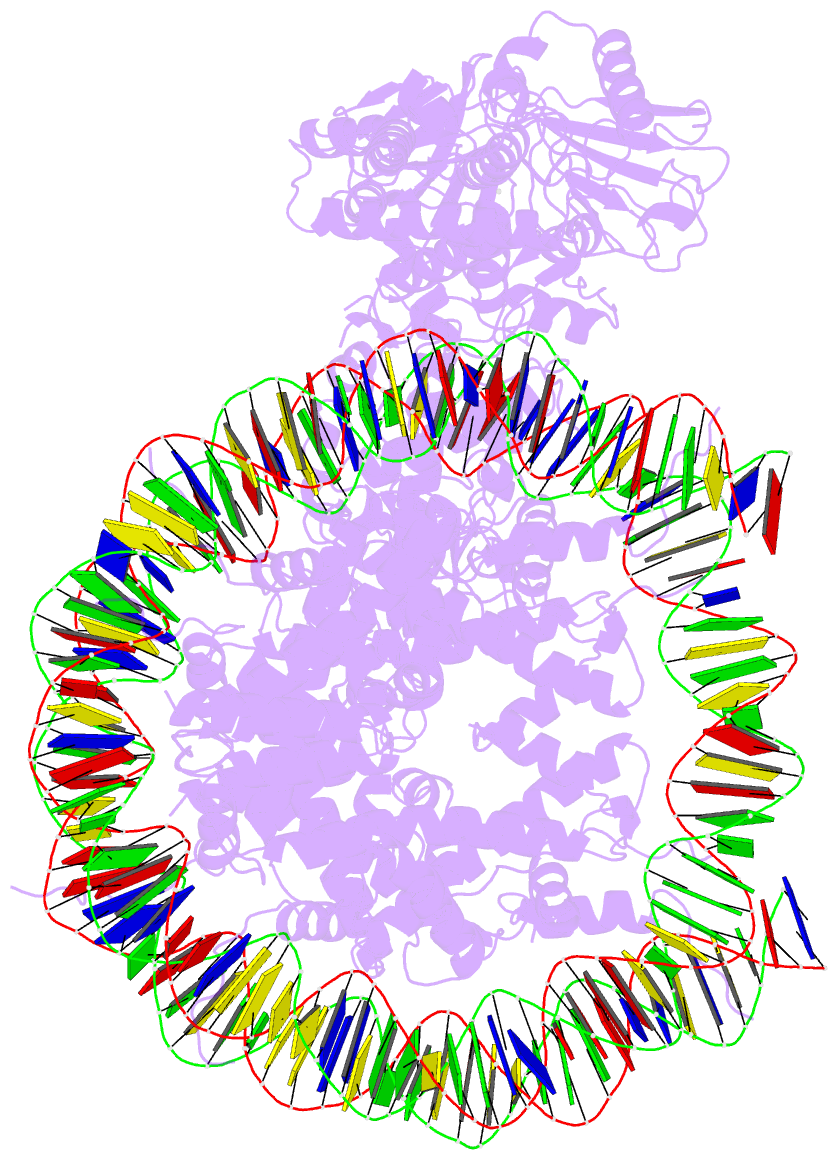
Summary information and primary citation
- PDB-id
- 6r25; DSSR-derived features in text and JSON formats
- Class
- gene regulation
- Method
- cryo-EM (4.61 Å)
- Summary
- Structure of lsd2-npac-linker-nucleosome core particle complex: class 3
- Reference
- Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A (2019): "A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex." Cell Rep, 27, 387-399.e7. doi: 10.1016/j.celrep.2019.03.061.
- Abstract
- LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes.