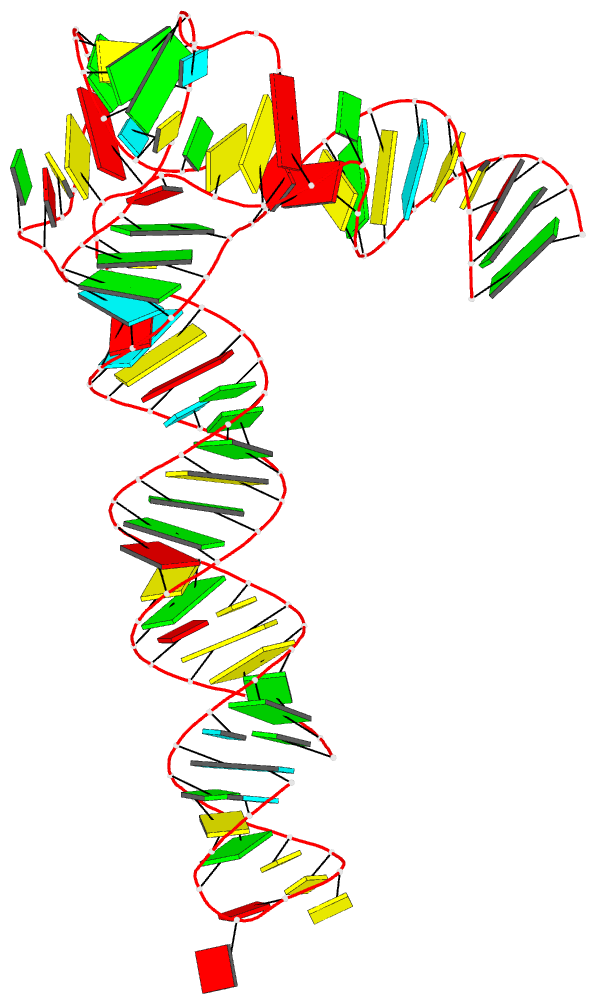
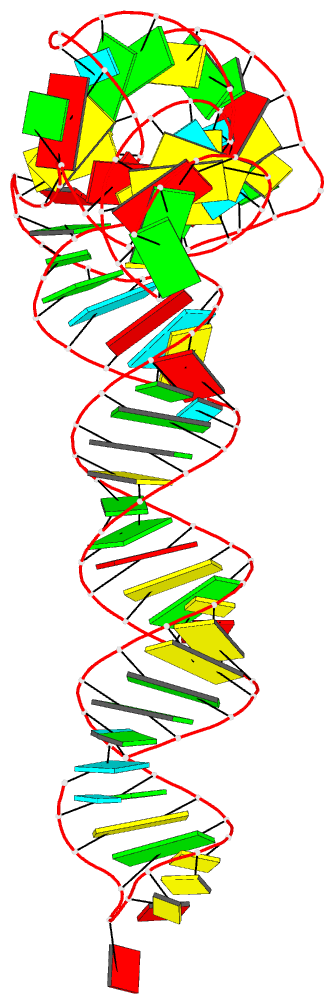
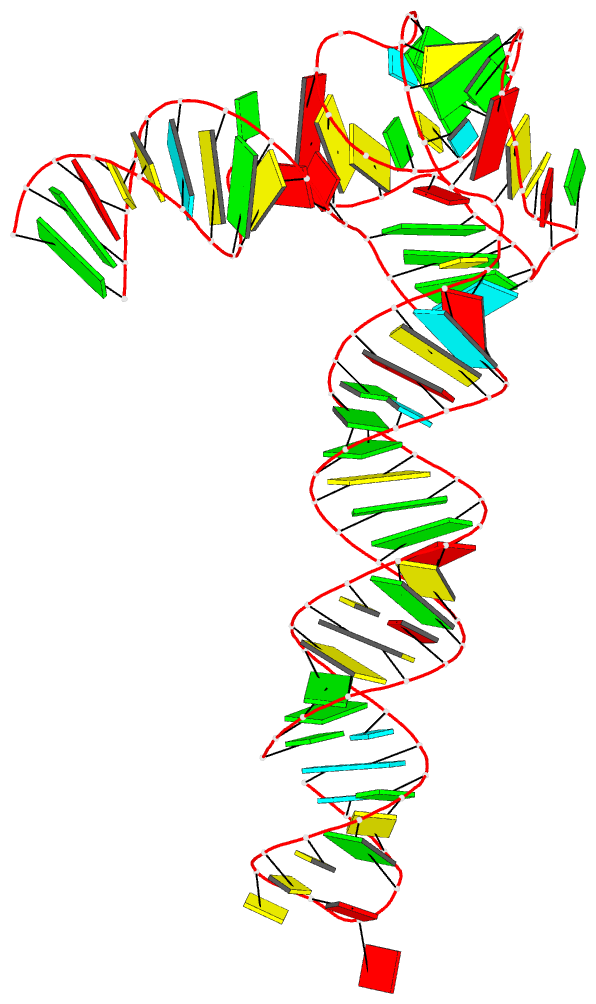
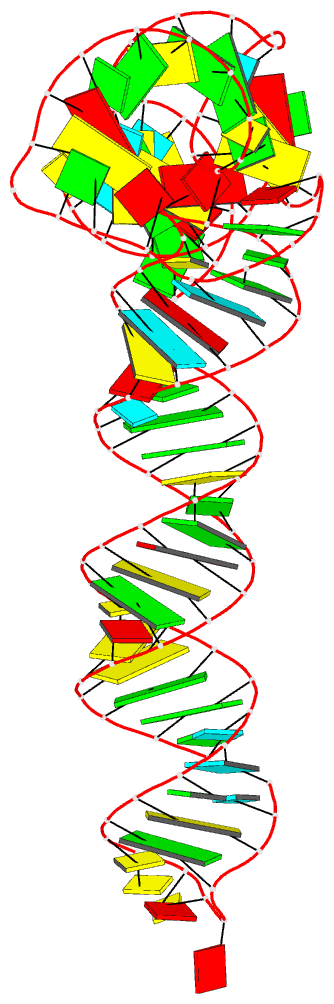
Summary information and primary citation
- PDB-id
-
6ol3;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (2.74 Å)
- Summary
- Crystal structure of an adenovirus virus-associated
RNA
- Reference
-
Hood IV, Gordon JM, Bou-Nader C, Henderson FE, Bahmanjah
S, Zhang J (2019): "Crystal
structure of an adenovirus virus-associated RNA."
Nat Commun, 10, 2871. doi:
10.1038/s41467-019-10752-6.
- Abstract
- Adenovirus Virus-Associated (VA) RNAs are the first
discovered viral noncoding RNAs. By mimicking
double-stranded RNAs (dsRNAs), the exceptionally abundant,
multifunctional VA RNAs sabotage host machineries that
sense, transport, process, or edit dsRNAs. How VA-I
suppresses PKR activation despite its strong dsRNA
character, and inhibits the crucial antiviral kinase to
promote viral translation, remains largely unknown. Here,
we report a 2.7 Å crystal structure of VA-I RNA. The
acutely bent VA-I features an unusually structured apical
loop, a wobble-enriched, coaxially stacked apical and
tetra-stems necessary and sufficient for PKR inhibition,
and a central domain pseudoknot that resembles
codon-anticodon interactions and prevents PKR activation by
VA-I. These global and local structural features
collectively define VA-I as an archetypal PKR inhibitor
made of RNA. The study provides molecular insights into how
viruses circumnavigate cellular rules of self vs non-self
RNAs to not only escape, but further compromise host innate
immunity.