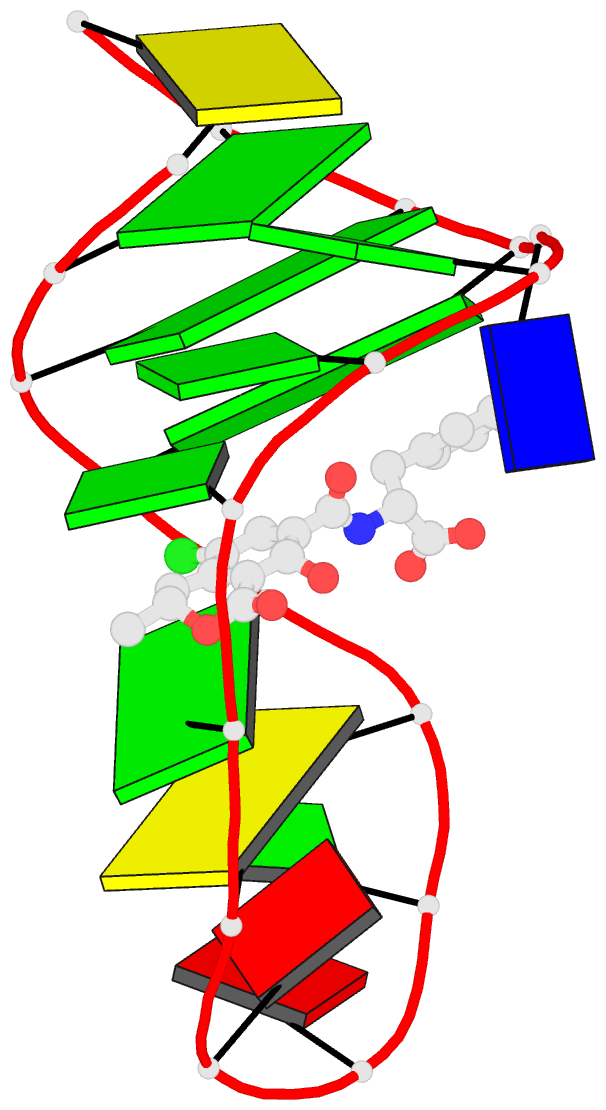
Summary information and primary citation
- PDB-id
- 6j2w; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- NMR
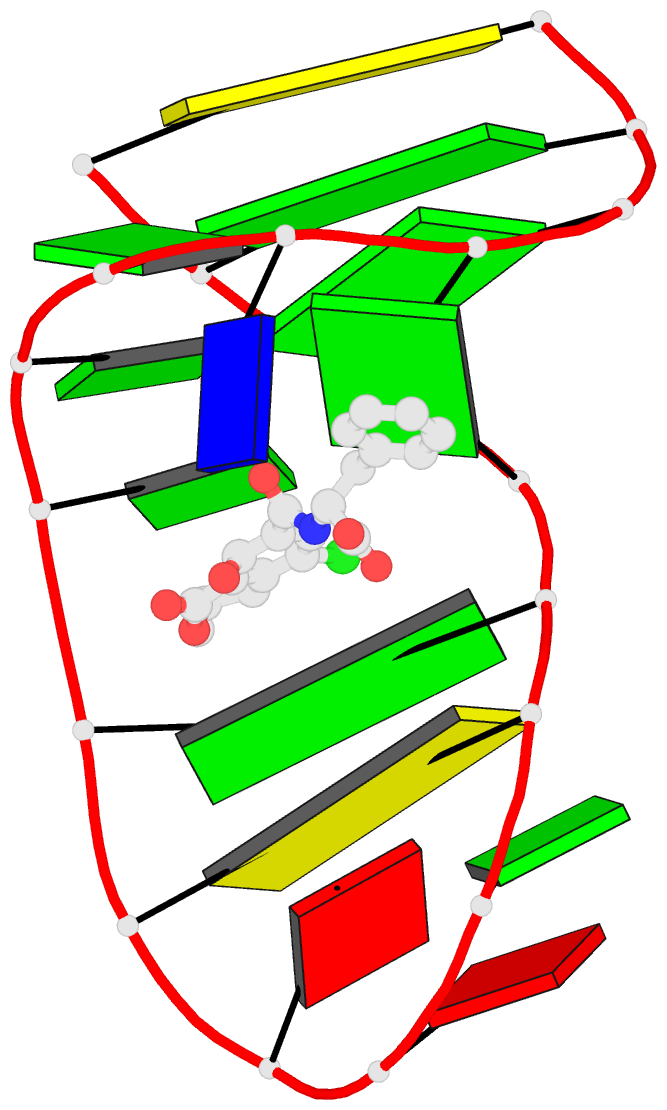
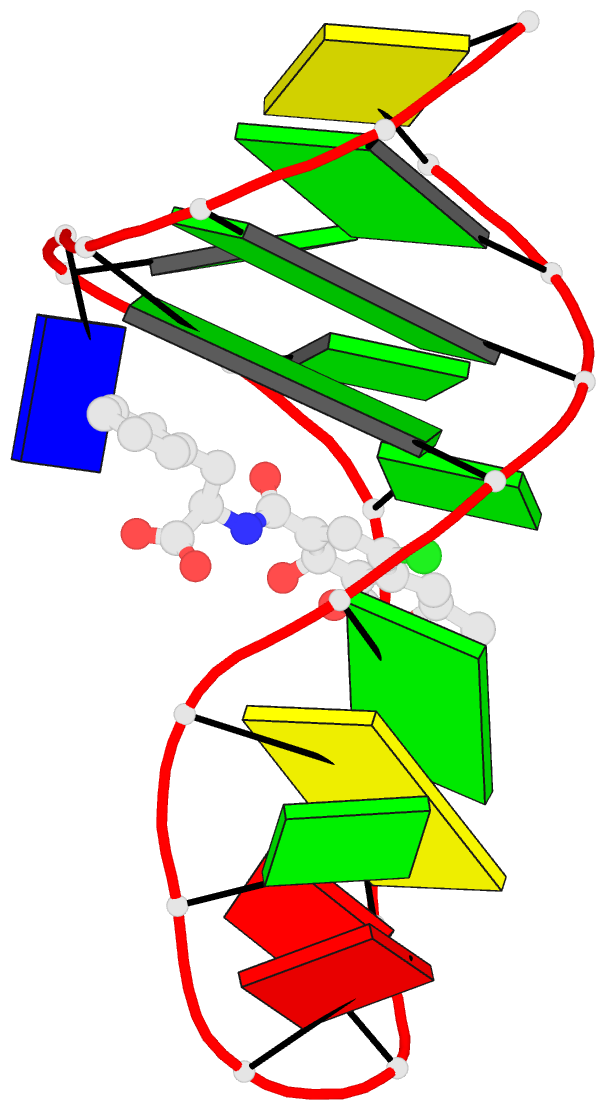
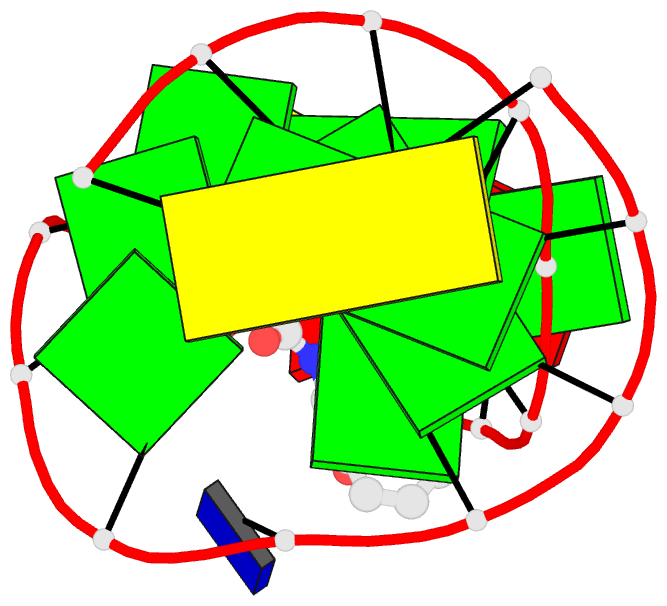
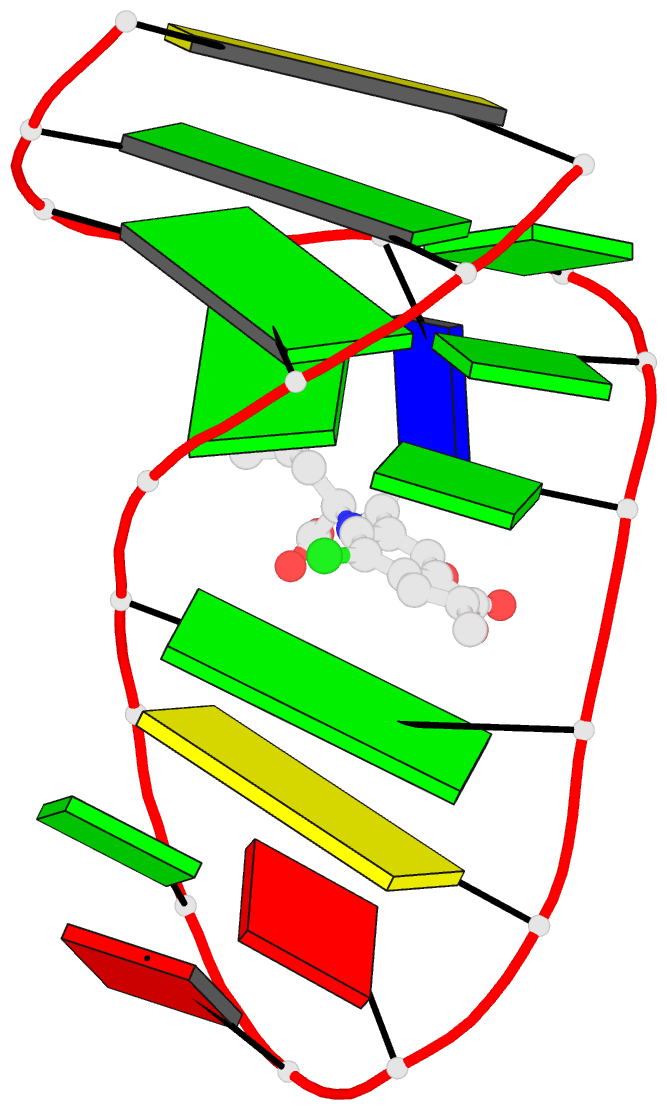
- Summary
- The structure of oba3-ota complex
- Reference
- Xu G, Zhao J, Liu N, Yang M, Zhao Q, Li C, Liu M (2019): "Structure-guided post-SELEX optimization of an ochratoxin A aptamer." Nucleic Acids Res., 47, 5963-5972. doi: 10.1093/nar/gkz336.
- Abstract
- SELEX is the cornerstone for aptamer research with broad applications in biosensors and medicine. To improve the affinity of selected aptamers, we propose a structure-guided post-SELEX approach, an optimization method based on the precise secondary structure of the aptamer-ligand complex. We demonstrate this approach using the Ochratoxin A (OTA) aptamer. Guided by the structure, we designed a new aptamer whose affinity is improved by more than 50-fold. We also determined the high-resolution NMR structure of the new aptamer-OTA complex and elucidated the discriminatory recognition mechanism of one atomic difference between two analogs, OTA and OTB. The aptamer forms an unusual hairpin structure containing an intramolecular triple helix, which is not seen in the previously determined aptamer complex. The π-π stacking, the hydrophobic interaction, hydrogen bonds and halogen bonds between OTA and the aptamer contribute to the recognition of OTA, and the halogen bonds play an important role in discriminating between OTA and OTB. Our results demonstrate that the structure-guided post-SELEX approach improves aptamers affinity. An improved OTA biosensor system might be developed using this new strategy.