Summary information and primary citation
- PDB-id
-
6gzr;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
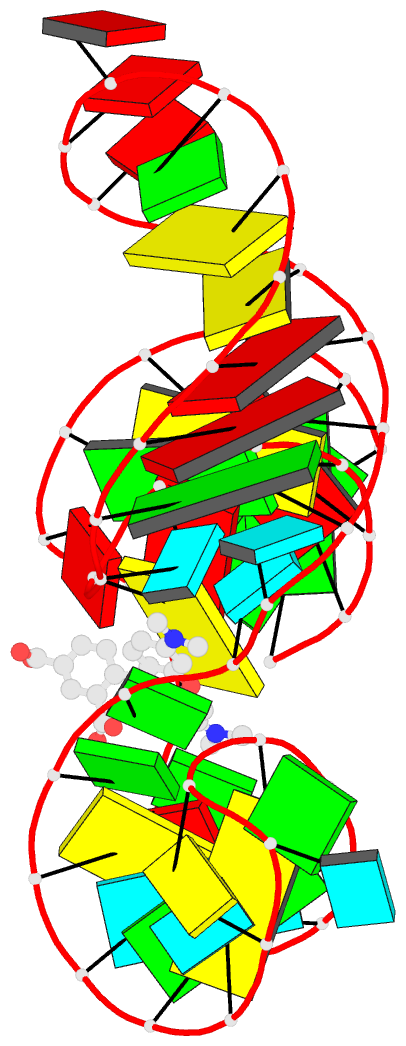
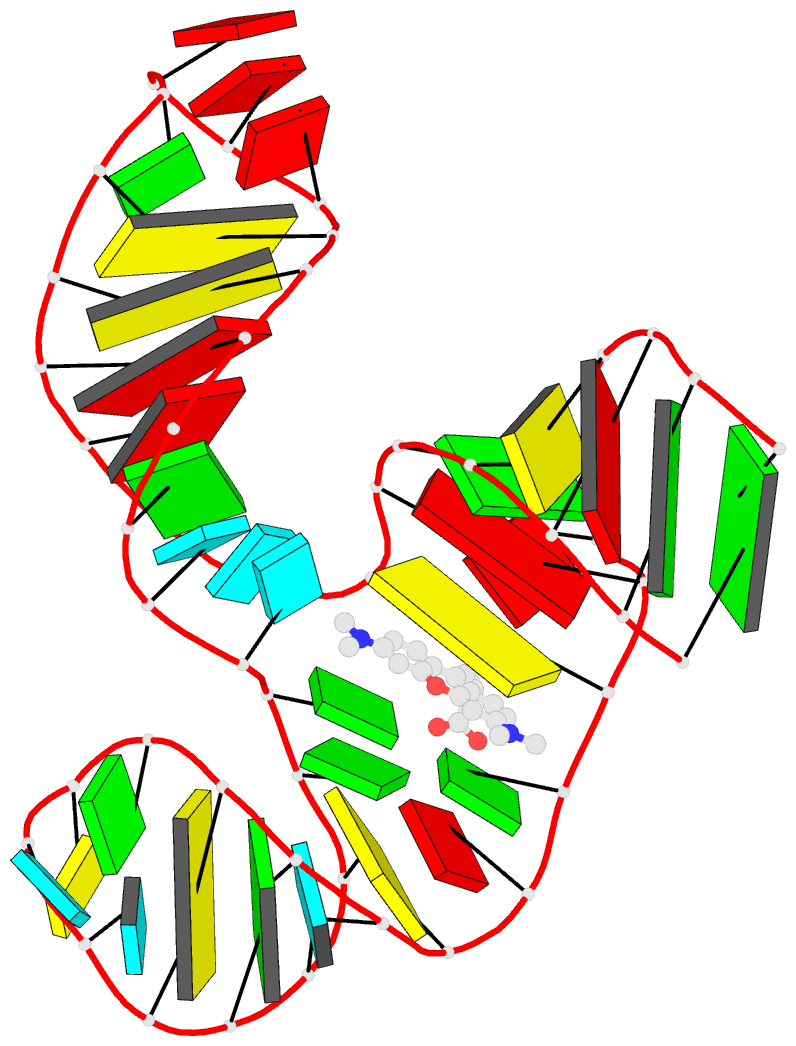
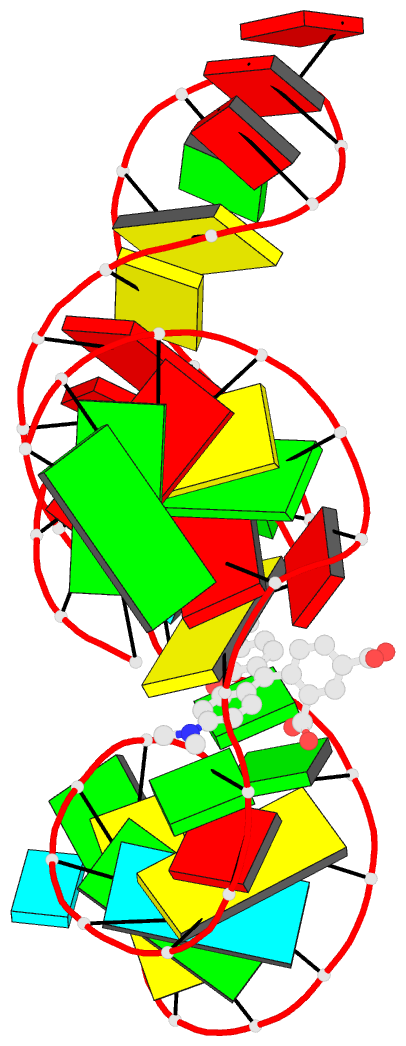
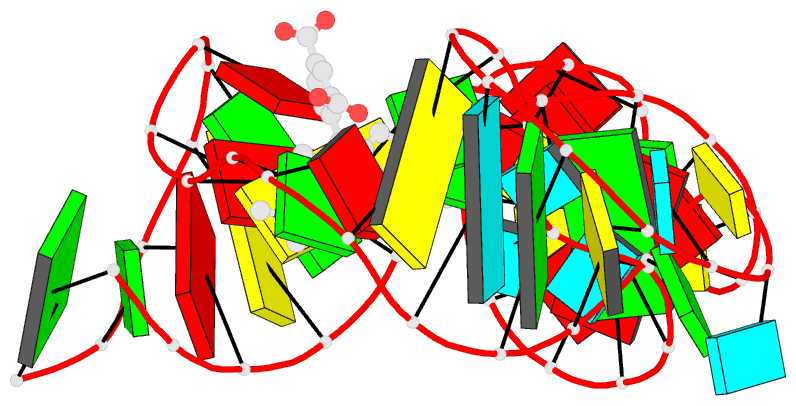
- Solution NMR structure of the tetramethylrhodamine
(tmr) aptamer 3 in complex with 5-tamra
- Reference
-
Duchardt-Ferner E, Juen M, Bourgeois B, Madl T, Kreutz C,
Ohlenschlager O, Wohnert J (2020): "Structure
of an RNA aptamer in complex with the fluorophore
tetramethylrhodamine." Nucleic Acids Res.,
48, 949-961. doi: 10.1093/nar/gkz1113.
- Abstract
- RNA aptamers-artificially created RNAs with high
affinity and selectivity for their target ligand generated
from random sequence pools-are versatile tools in the
fields of biotechnology and medicine. On a more fundamental
level, they also further our general understanding of
RNA-ligand interactions e. g. in regard to the relationship
between structural complexity and ligand affinity and
specificity, RNA structure and RNA folding. Detailed
structural knowledge on a wide range of aptamer-ligand
complexes is required to further our understanding of
RNA-ligand interactions. Here, we present the atomic
resolution structure of an RNA-aptamer binding to the
fluorescent xanthene dye tetramethylrhodamine. The high
resolution structure, solved by NMR-spectroscopy in
solution, reveals binding features both common and
different from the binding mode of other aptamers with
affinity for ligands carrying planar aromatic ring systems
such as the malachite green aptamer which binds to the
tetramethylrhodamine related dye malachite green or the
flavin mononucleotide aptamer.