Summary information and primary citation
- PDB-id
- 6cy4; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (1.851 Å)
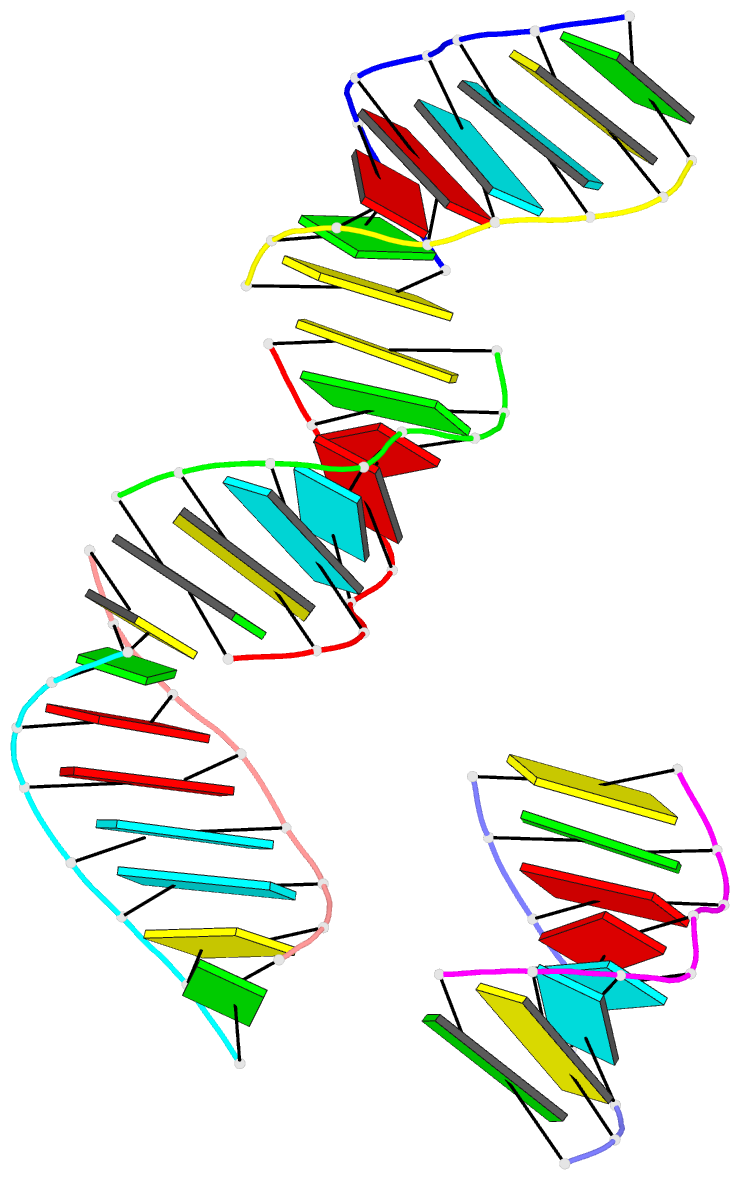
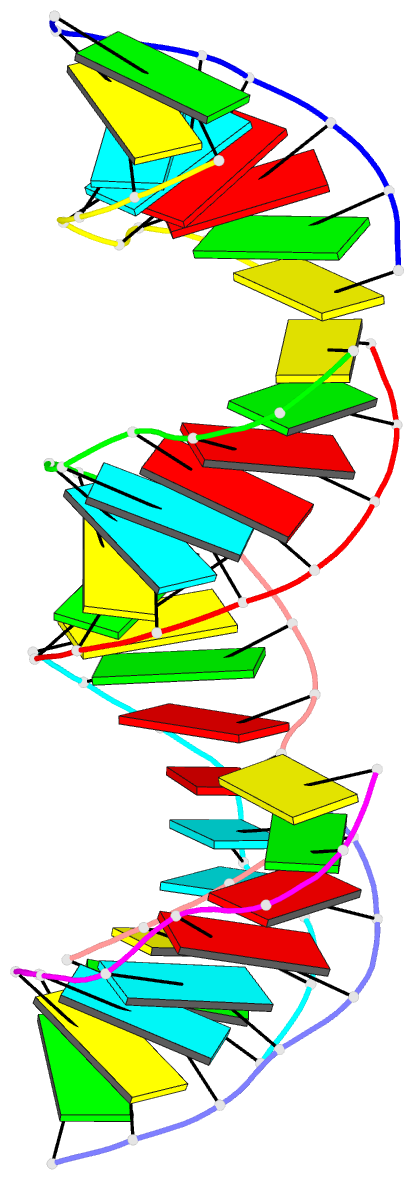
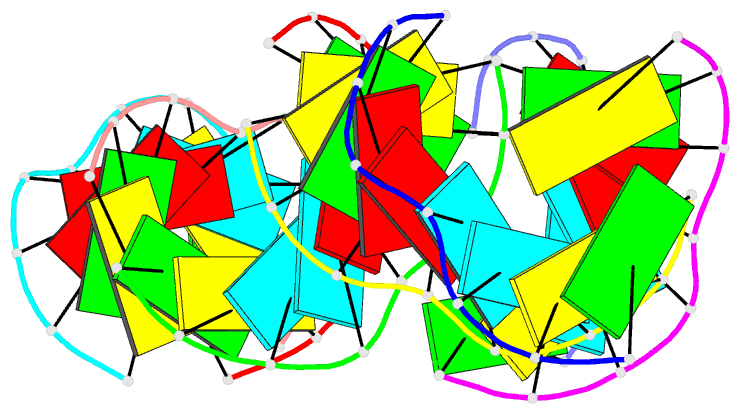
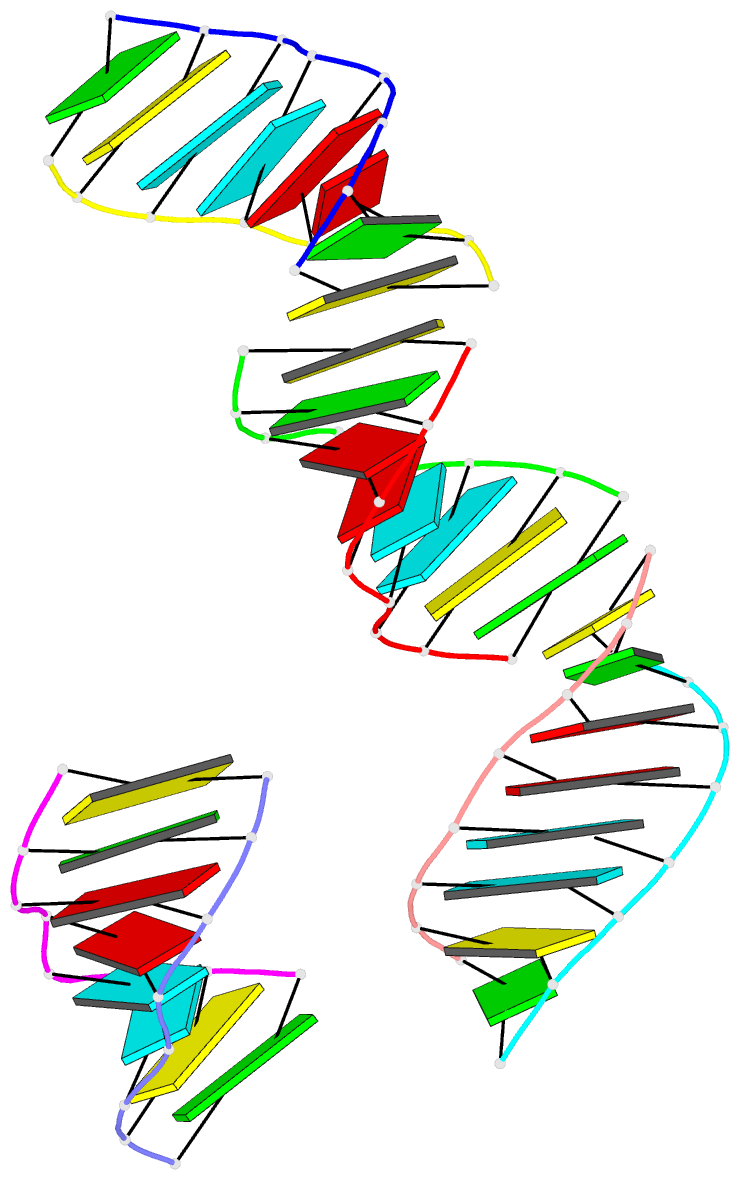
- Summary
- RNA octamer containing 2'-ome, 4'- cbeta-ome u.
- Reference
- Harp JM, Guenther DC, Bisbe A, Perkins L, Matsuda S, Bommineni GR, Zlatev I, Foster DJ, Taneja N, Charisse K, Maier MA, Rajeev KG, Manoharan M, Egli M (2018): "Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity." Nucleic Acids Res., 46, 8090-8104. doi: 10.1093/nar/gky703.
- Abstract
- Chemical modification is a prerequisite of oligonucleotide therapeutics for improved metabolic stability, uptake and activity, irrespective of their mode of action, i.e. antisense, RNAi or aptamer. Phosphate moiety and ribose C2'/O2' atoms are the most common sites for modification. Compared to 2'-O-substituents, ribose 4'-C-substituents lie in proximity of both the 3'- and 5'-adjacent phosphates. To investigate potentially beneficial effects on nuclease resistance we combined 2'-F and 2'-OMe with 4'-Cα- and 4'-Cβ-OMe, and 2'-F with 4'-Cα-methyl modification. The α- and β-epimers of 4'-C-OMe-uridine and the α-epimer of 4'-C-Me-uridine monomers were synthesized and incorporated into siRNAs. The 4'α-epimers affect thermal stability only minimally and show increased nuclease stability irrespective of the 2'-substituent (H, F, OMe). The 4'β-epimers are strongly destabilizing, but afford complete resistance against an exonuclease with the phosphate or phosphorothioate backbones. Crystal structures of RNA octamers containing 2'-F,4'-Cα-OMe-U, 2'-F,4'-Cβ-OMe-U, 2'-OMe,4'-Cα-OMe-U, 2'-OMe,4'-Cβ-OMe-U or 2'-F,4'-Cα-Me-U help rationalize these observations and point to steric and electrostatic origins of the unprecedented nuclease resistance seen with the chain-inverted 4'β-U epimer. We used structural models of human Argonaute 2 in complex with guide siRNA featuring 2'-F,4'-Cα-OMe-U or 2'-F,4'-Cβ-OMe-U at various sites in the seed region to interpret in vitro activities of siRNAs with the corresponding 2'-/4'-C-modifications.