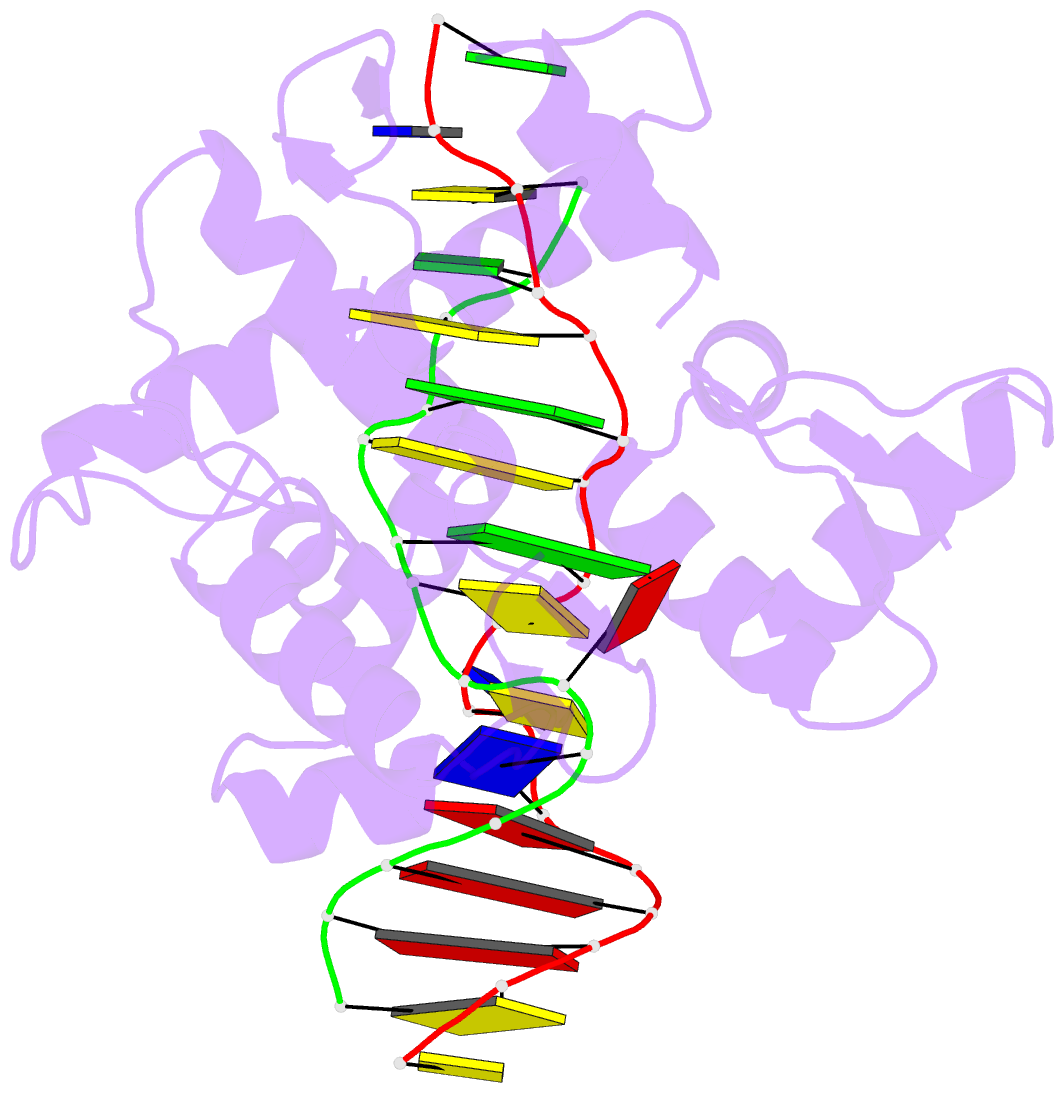
Summary information and primary citation
- PDB-id
- 5zu1; DSSR-derived features in text and JSON formats
- Class
- hydrolase-DNA
- Method
- X-ray (3.009 Å)
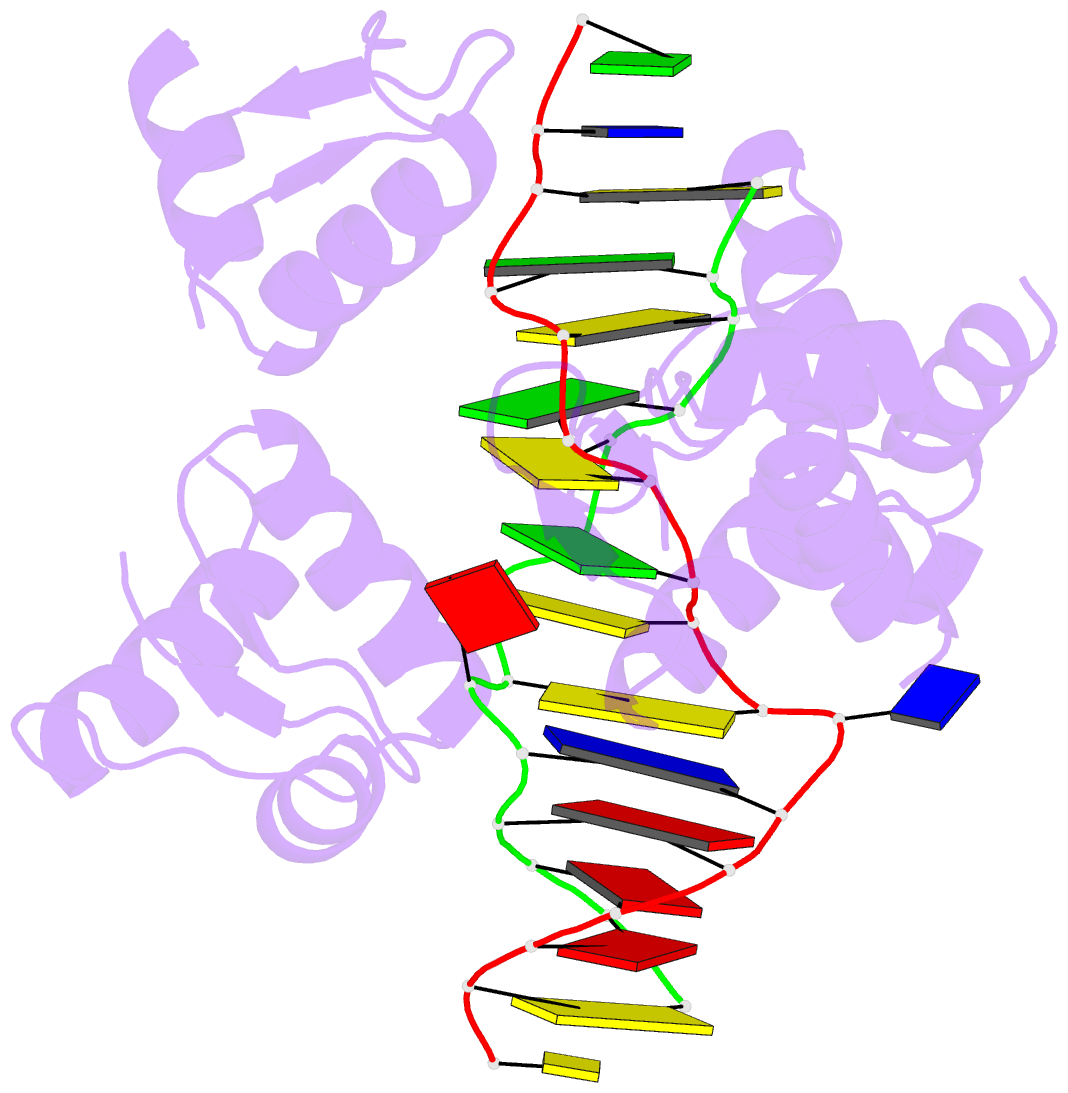
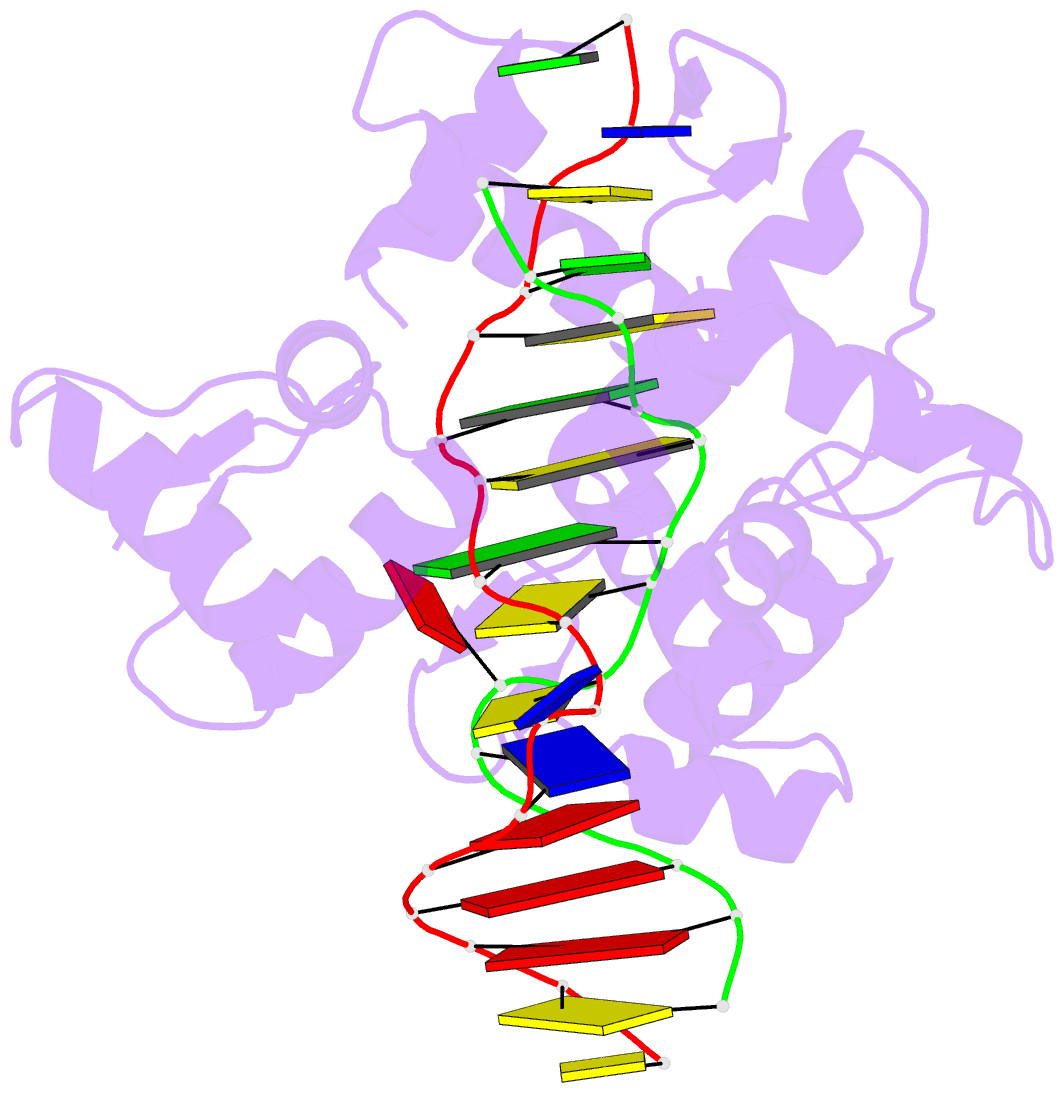
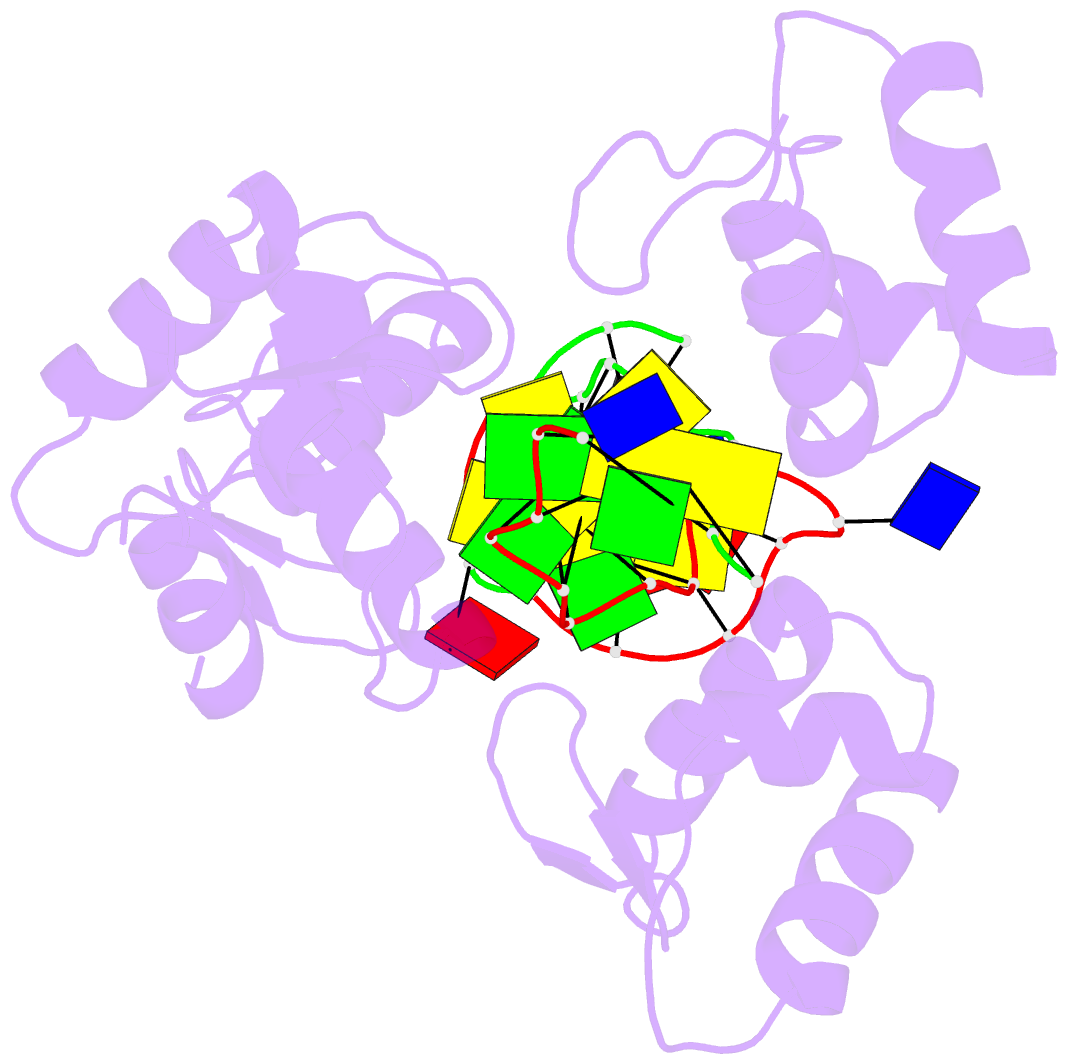
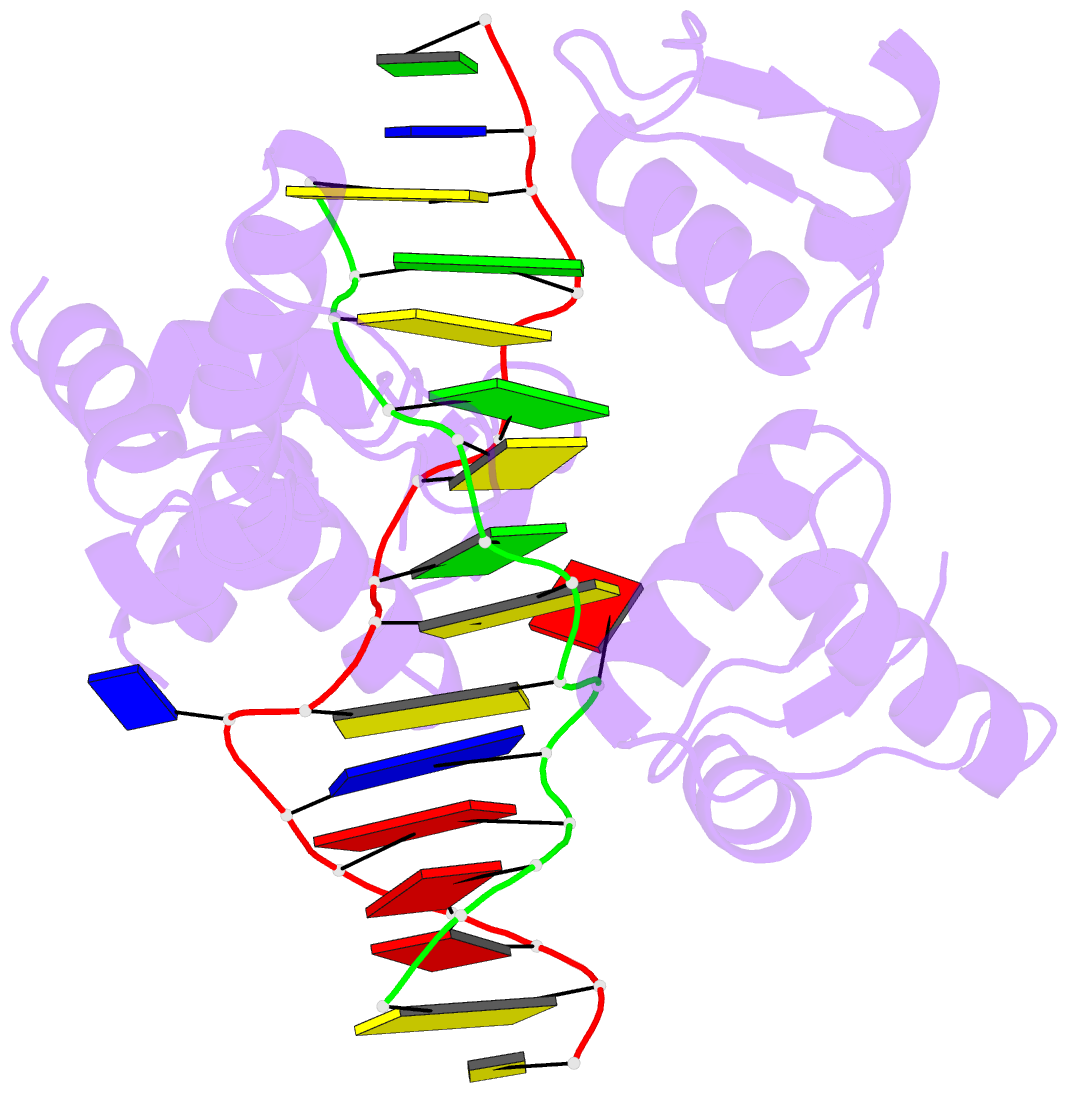
- Summary
- Crystal structure of bz junction in diverse sequence
- Reference
- Kim D, Hur J, Han JH, Ha SC, Shin D, Lee S, Park S, Sugiyama H, Kim KK (2018): "Sequence preference and structural heterogeneity of BZ junctions." Nucleic Acids Res., 46, 10504-10513. doi: 10.1093/nar/gky784.
- Abstract
- BZ junctions, which connect B-DNA to Z-DNA, are necessary for local transformation of B-DNA to Z-DNA in the genome. However, the limited information on the junction-forming sequences and junction structures has led to a lack of understanding of the structural diversity and sequence preferences of BZ junctions. We determined three crystal structures of BZ junctions with diverse sequences followed by spectroscopic validation of DNA conformation. The structural features of the BZ junctions were well conserved regardless of sequences via the continuous base stacking through B-to-Z DNA with A-T base extrusion. However, the sequence-dependent structural heterogeneity of the junctions was also observed in base step parameters that are correlated with steric constraints imposed during Z-DNA formation. Further, circular dichroism and fluorescence-based analysis of BZ junctions revealed that a base extrusion was only found at the A-T base pair present next to a stable dinucleotide Z-DNA unit. Our findings suggest that Z-DNA formation in the genome is influenced by the sequence preference for BZ junctions.