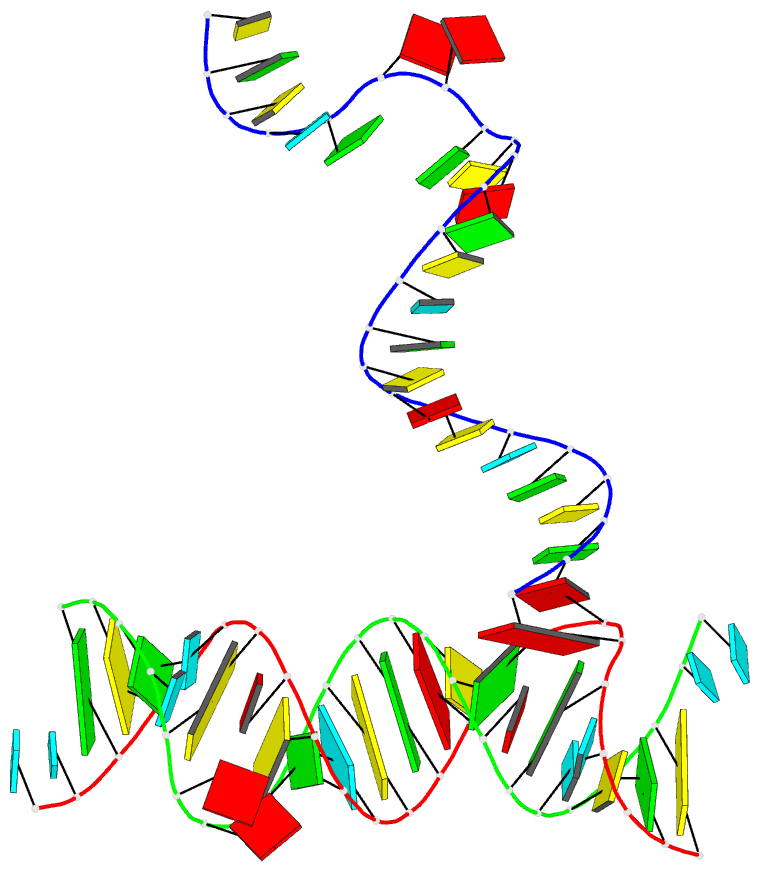
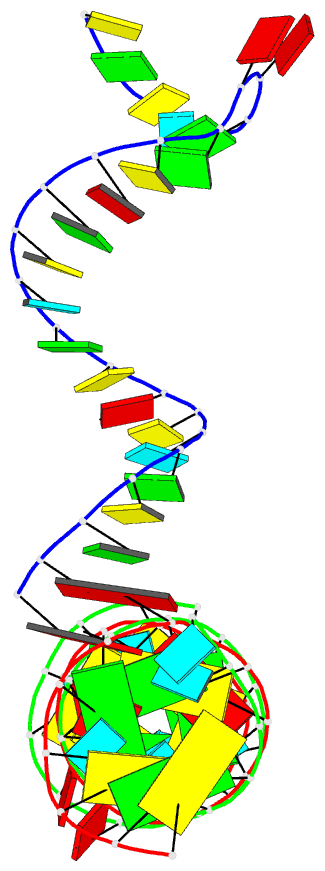
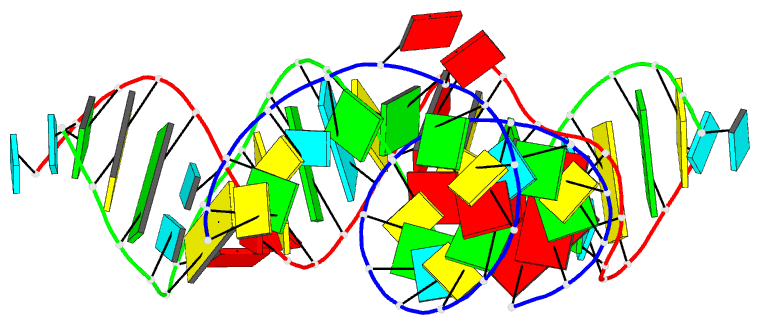
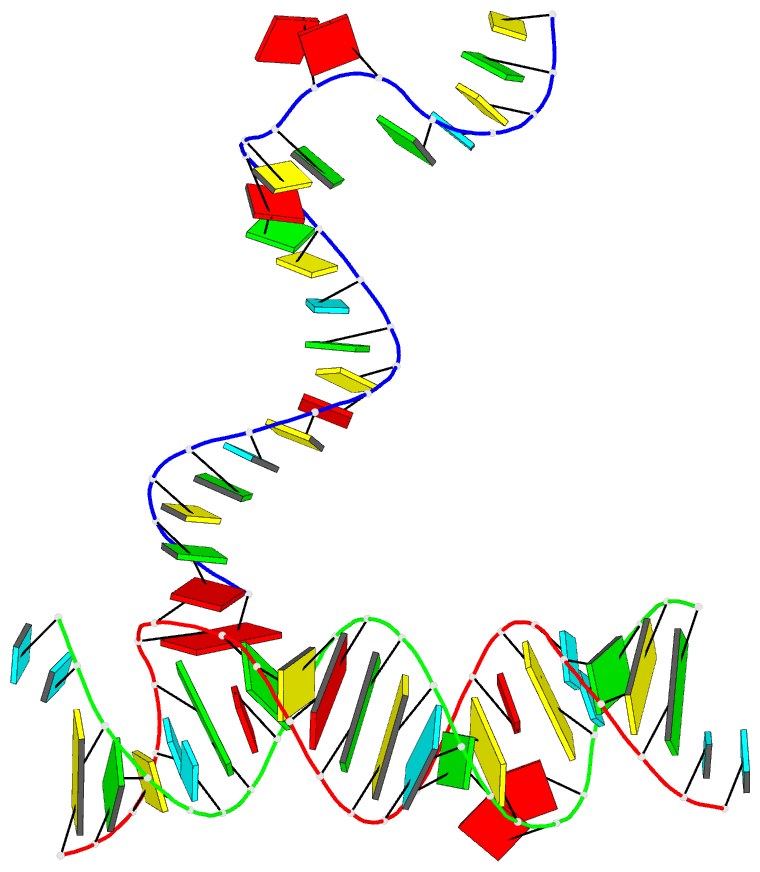
Summary information and primary citation
- PDB-id
- 5zeg; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.4 Å)
- Summary
- Crystal structure of the bacterial a1408me1a-mutant ribosomal decoding site
- Reference
- Kanazawa H, Baba F, Koganei M, Kondo J (2017): "A structural basis for the antibiotic resistance conferred by an N1-methylation of A1408 in 16S rRNA." Nucleic Acids Res., 45, 12529-12535. doi: 10.1093/nar/gkx882.
- Abstract
- The aminoglycoside resistance conferred by an N1-methylation of A1408 in 16S rRNA by a novel plasmid-mediated methyltransferase NpmA can be a future health threat. In the present study, we have determined crystal structures of the bacterial ribosomal decoding A site with an A1408m1A antibiotic-resistance mutation both in the presence and absence of aminoglycosides. G418 and paromomycin both possessing a 6'-OH group specifically bind to the mutant A site and disturb its function as a molecular switch in the decoding process. On the other hand, binding of gentamicin with a 6'-NH3+ group to the mutant A site could not be observed in the present crystal structure. These observations agree with the minimum inhibitory concentration of aminoglycosides against Escherichia coli. In addition, one of our crystal structures suggests a possible conformational change of A1408 during the N1-methylation reaction by NpmA. The structural information obtained explains how bacteria acquire resistance against aminoglycosides along with a minimum of fitness cost by the N1-methylation of A1408 and provides novel information for designing the next-generation aminoglycoside.