Summary information and primary citation
- PDB-id
- 5wsp; DSSR-derived features in text and JSON formats
- Class
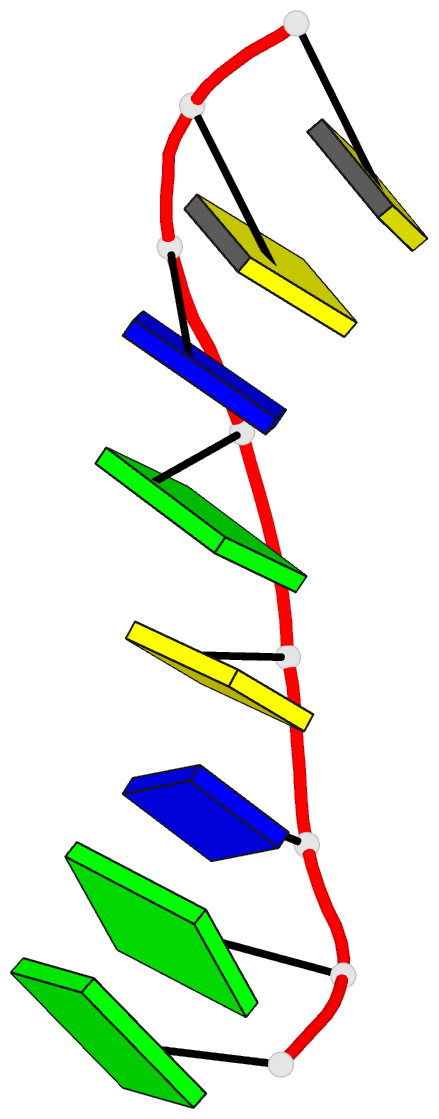
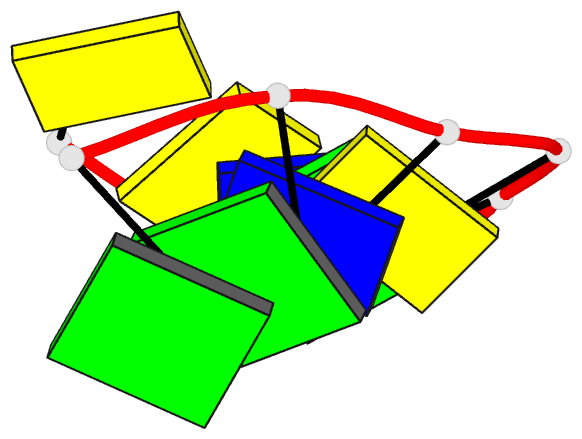
- DNA
- Method
- X-ray (1.502 Å)
- Summary
- Crystal structure of dna3 duplex
- Reference
- Liu HH, Cai C, Haruehanroengra P, Yao QQ, Chen YQ, Yang C, Luo Q, Wu BX, Li JX, Ma JB, Sheng J, Gan JH (2017): "Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex." Nucleic Acids Res., 45, 2910-2918. doi: 10.1093/nar/gkw1296.
- Abstract
- Owing to their great potentials in genetic code extension and the development of nucleic acid-based functional nanodevices, DNA duplexes containing Hg(II)-mediated base pairs have been extensively studied during the past 60 years. However, structural basis underlying these base pairs remains poorly understood. Herein, we present five high-resolution crystal structures including one first-time reported C-Hg(II)-T containing duplex, three T-Hg(II)-T containing duplexes and one native duplex containing T-T pair without Hg(II) Our structures suggest that both C-T and T-T pairs are flexible in interacting with the Hg(II) ion with various binding modes including N3-Hg(II)-N3, N4-Hg(II)-N3, O2-Hg(II)-N3 and N3-Hg(II)-O4. Our studies also reveal that the overall conformations of the C-Hg(II)-T and T-Hg(II)-T pairs are affected by their neighboring residues via the interactions with the solvent molecules or other metal ions, such as Sr(II) These results provide detailed insights into the interactions between Hg(II) and nucleobases and the structural basis for the rational design of C-Hg(II)-T or T-Hg(II)-T containing DNA nanodevices in the future.