Summary information and primary citation
- PDB-id
- 5wnr; DSSR-derived features in text and JSON formats
- Class
- ribosome
- Method
- X-ray (3.5 Å)
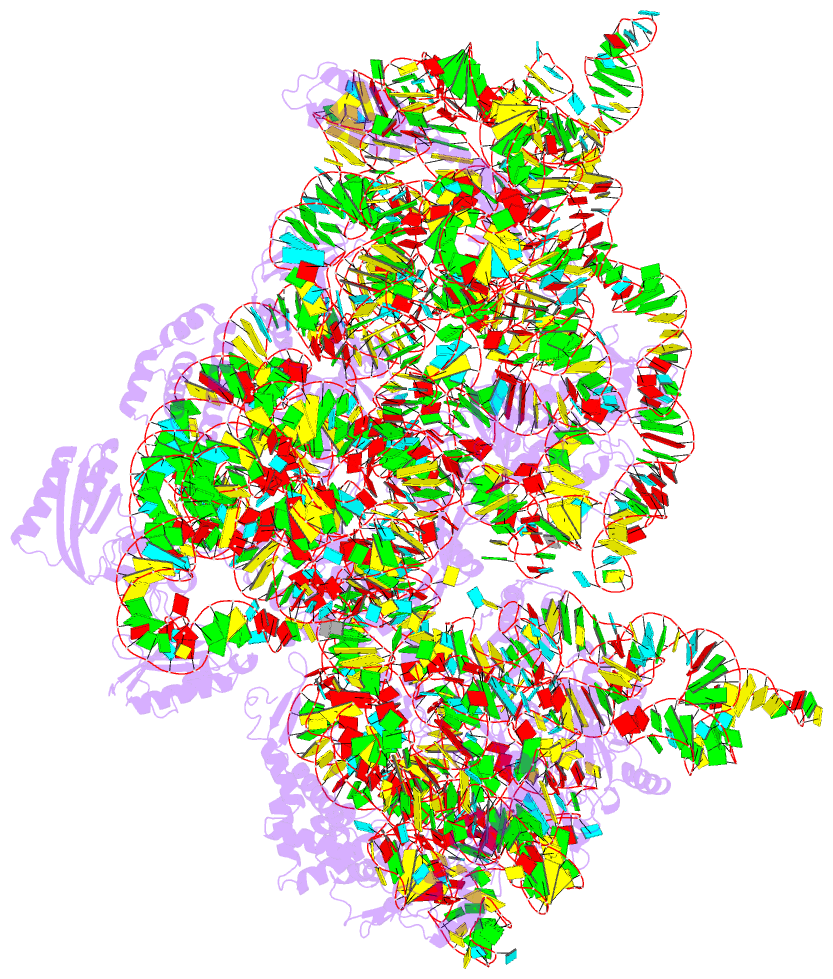
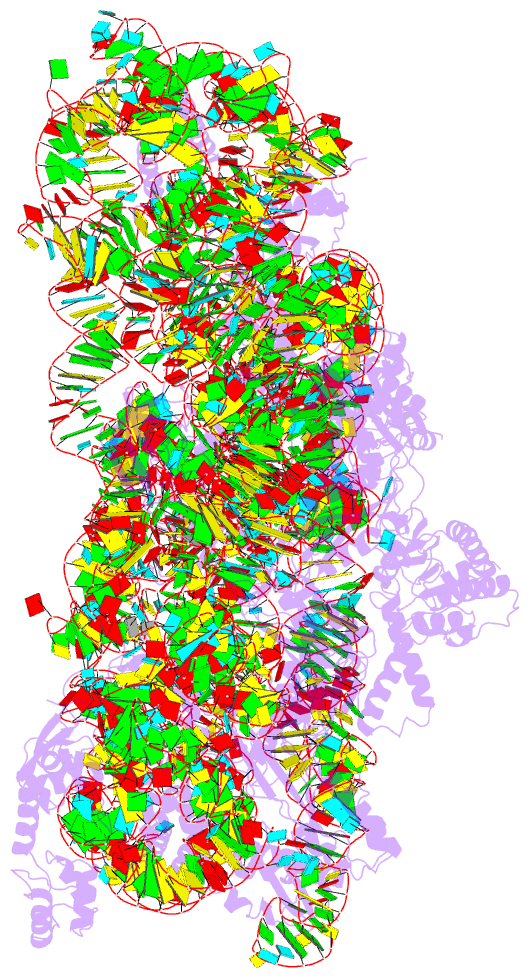
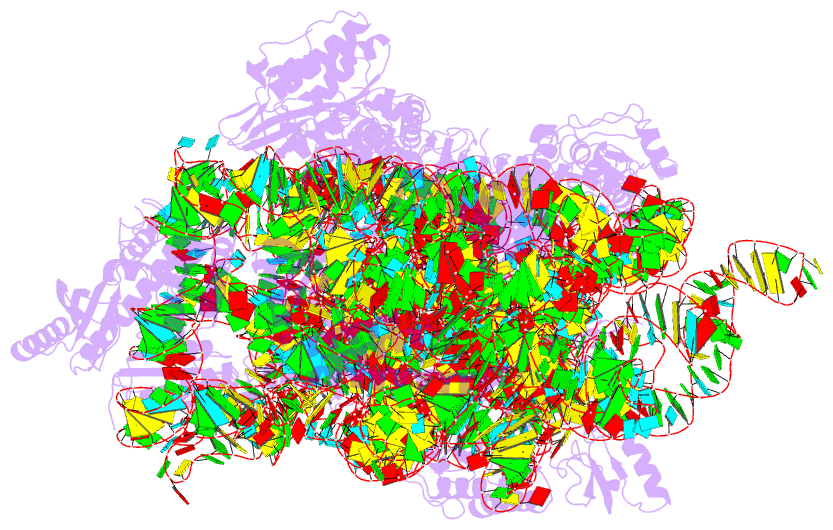
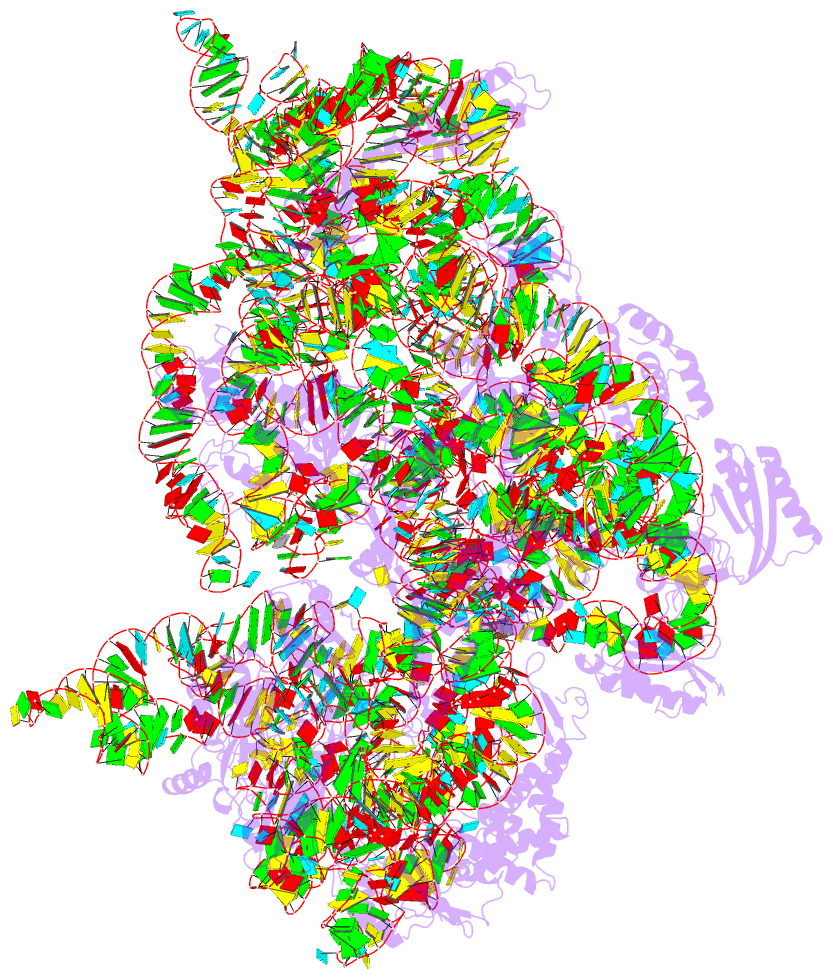
- Summary
- Crystal structure of 30s ribosomal subunit from thermus thermophilus
- Reference
- Choi J, Indrisiunaite G, DeMirci H, Ieong KW, Wang J, Petrov A, Prabhakar A, Rechavi G, Dominissini D, He C, Ehrenberg M, Puglisi JD (2018): "2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation." Nat. Struct. Mol. Biol., 25, 208-216. doi: 10.1038/s41594-018-0030-z.
- Abstract
- Chemical modifications of mRNA may regulate many aspects of mRNA processing and protein synthesis. Recently, 2'-O-methylation of nucleotides was identified as a frequent modification in translated regions of human mRNA, showing enrichment in codons for certain amino acids. Here, using single-molecule, bulk kinetics and structural methods, we show that 2'-O-methylation within coding regions of mRNA disrupts key steps in codon reading during cognate tRNA selection. Our results suggest that 2'-O-methylation sterically perturbs interactions of ribosomal-monitoring bases (G530, A1492 and A1493) with cognate codon-anticodon helices, thereby inhibiting downstream GTP hydrolysis by elongation factor Tu (EF-Tu) and A-site tRNA accommodation, leading to excessive rejection of cognate aminoacylated tRNAs in initial selection and proofreading. Our current and prior findings highlight how chemical modifications of mRNA tune the dynamics of protein synthesis at different steps of translation elongation.