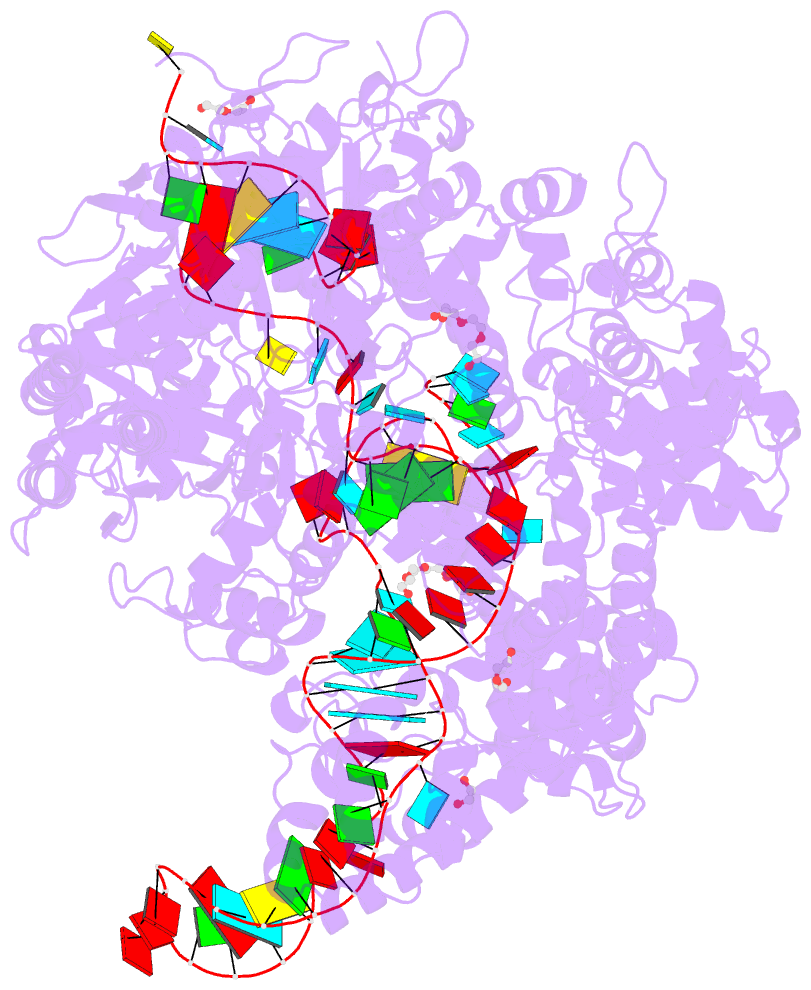
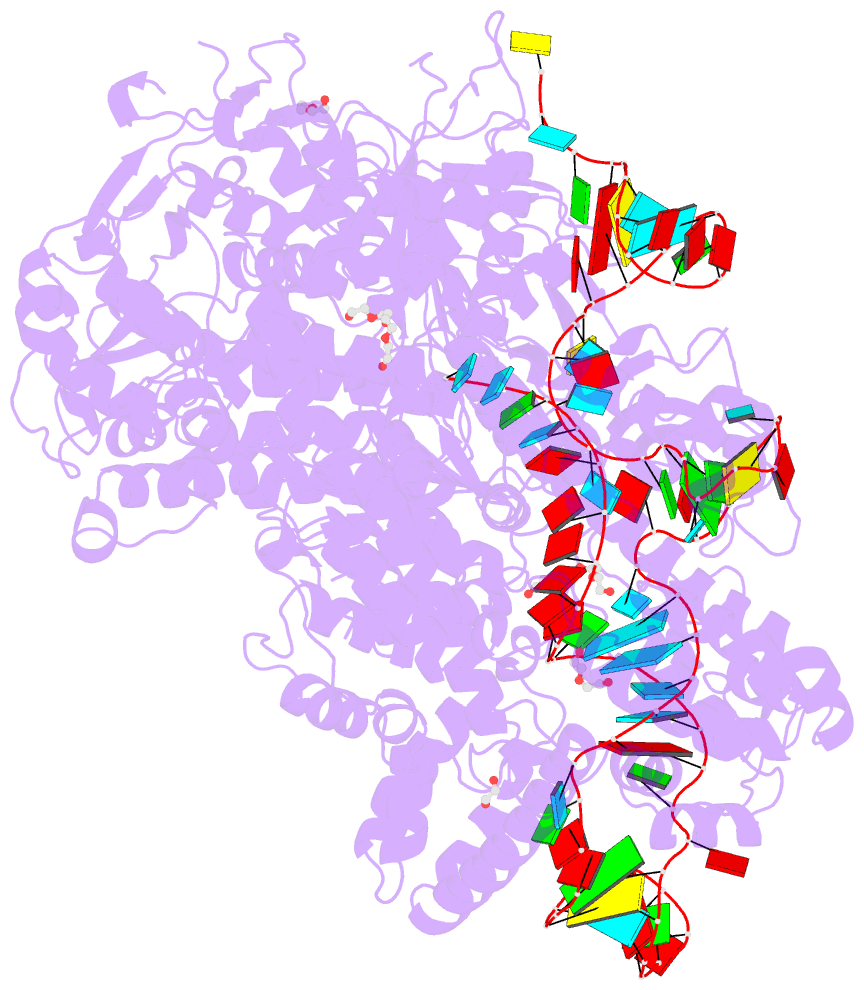
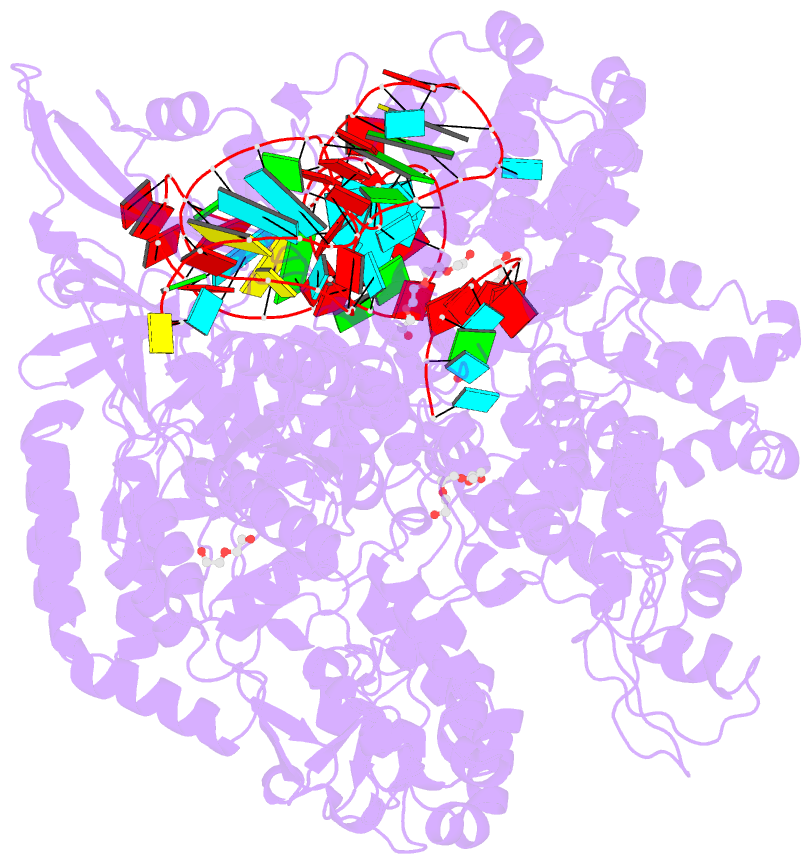
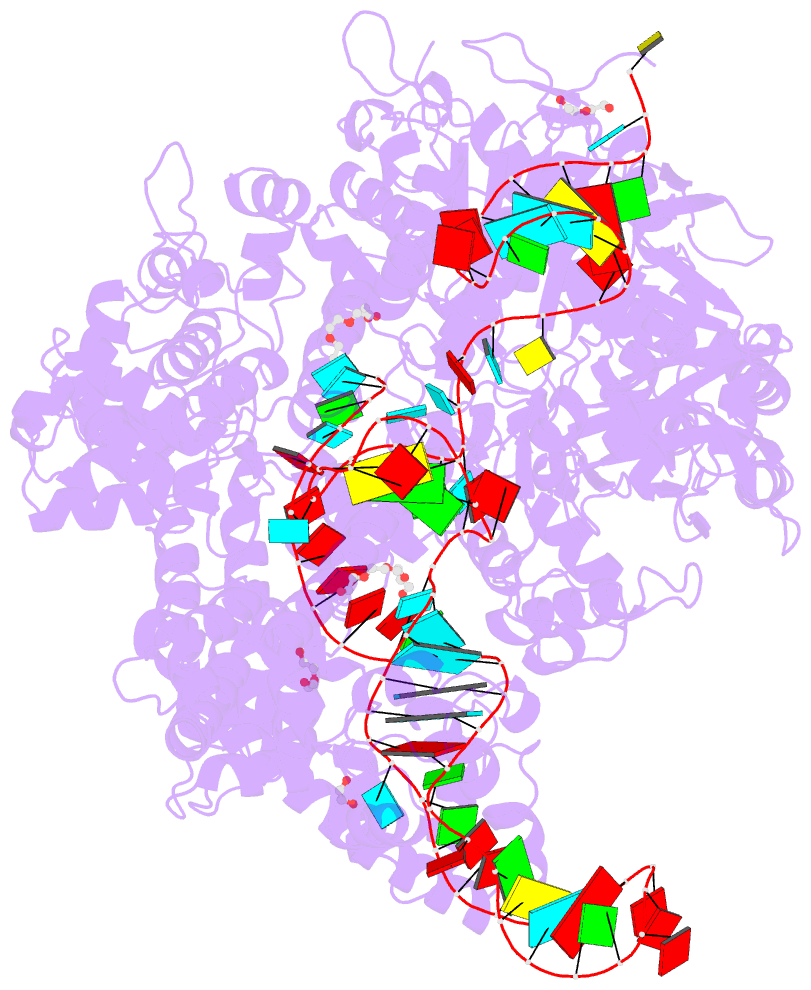
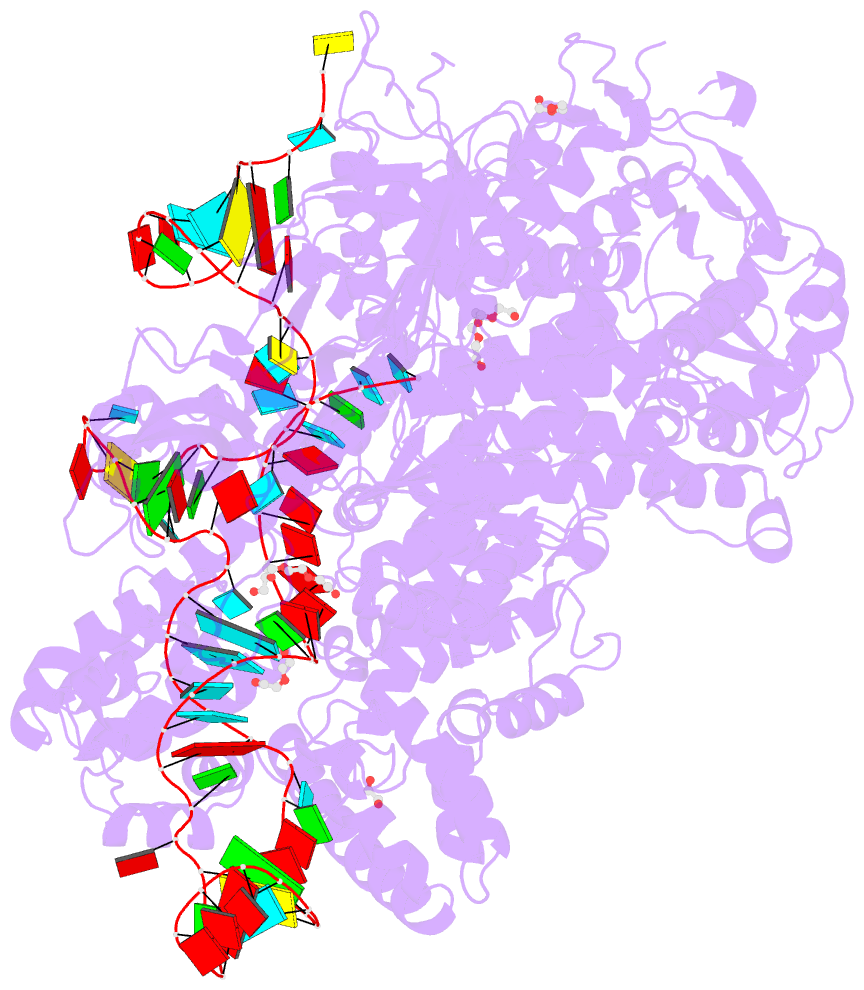
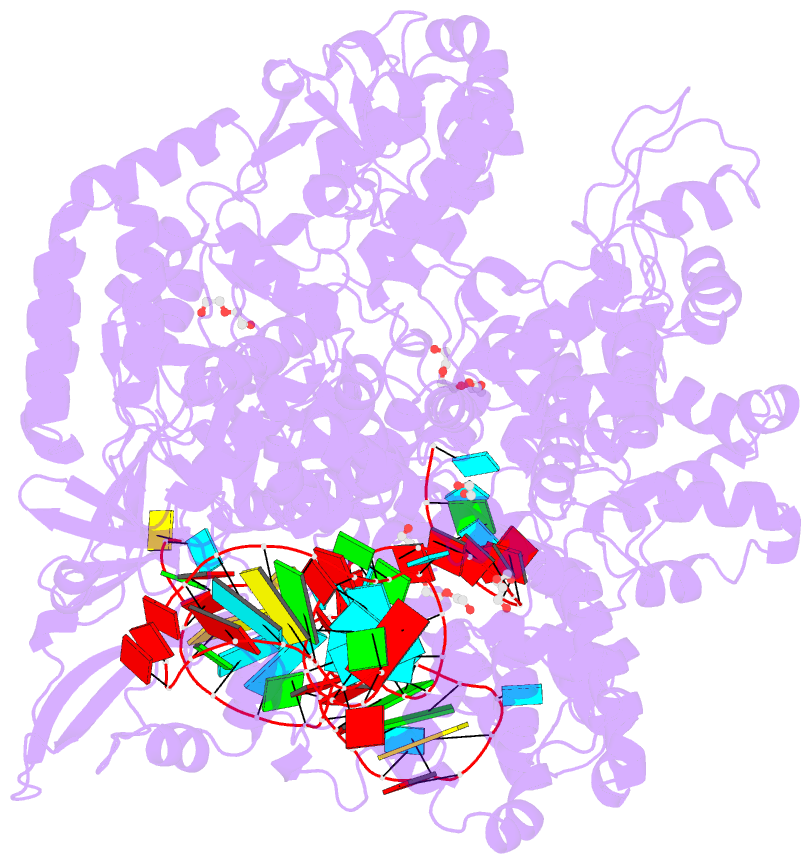
Summary information and primary citation
- PDB-id
- 5vw1; DSSR-derived features in text and JSON formats
- Class
- hydrolase
- Method
- X-ray (2.598 Å)
- Summary
- Crystal structure of spycas9-sgrna-acriia4 ternary complex
- Reference
- Yang H, Patel DJ (2017): "Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9." Mol. Cell, 67, 117-127.e5. doi: 10.1016/j.molcel.2017.05.024.
- Abstract
- Prokaryotic CRISPR-Cas adaptive immune systems utilize sequence-specific RNA-guided endonucleases to defend against infection by viruses, bacteriophages, and mobile elements, while these foreign genetic elements evolve diverse anti-CRISPR proteins to overcome the CRISPR-Cas-mediated defense of the host. Recently, AcrIIA2 and AcrIIA4, encoded by Listeria monocytogene prophages, were shown to block the endonuclease activity of type II-A Streptococcus pyogene Cas9 (SpyCas9). We now report the crystal structure of AcrIIA4 in complex with single-guide RNA-bound SpyCas9, thereby establishing that AcrIIA4 preferentially targets critical residues essential for PAM duplex recognition, as well as blocks target DNA access to key catalytic residues lining the RuvC pocket. These structural insights, validated by biochemical assays on key mutants, demonstrate that AcrIIA4 competitively occupies both PAM-interacting and non-target DNA strand cleavage catalytic pockets. Our studies provide insights into anti-CRISPR-mediated suppression mechanisms for inactivating SpyCas9, thereby broadening the applicability of CRISPR-Cas regulatory tools for genome editing.