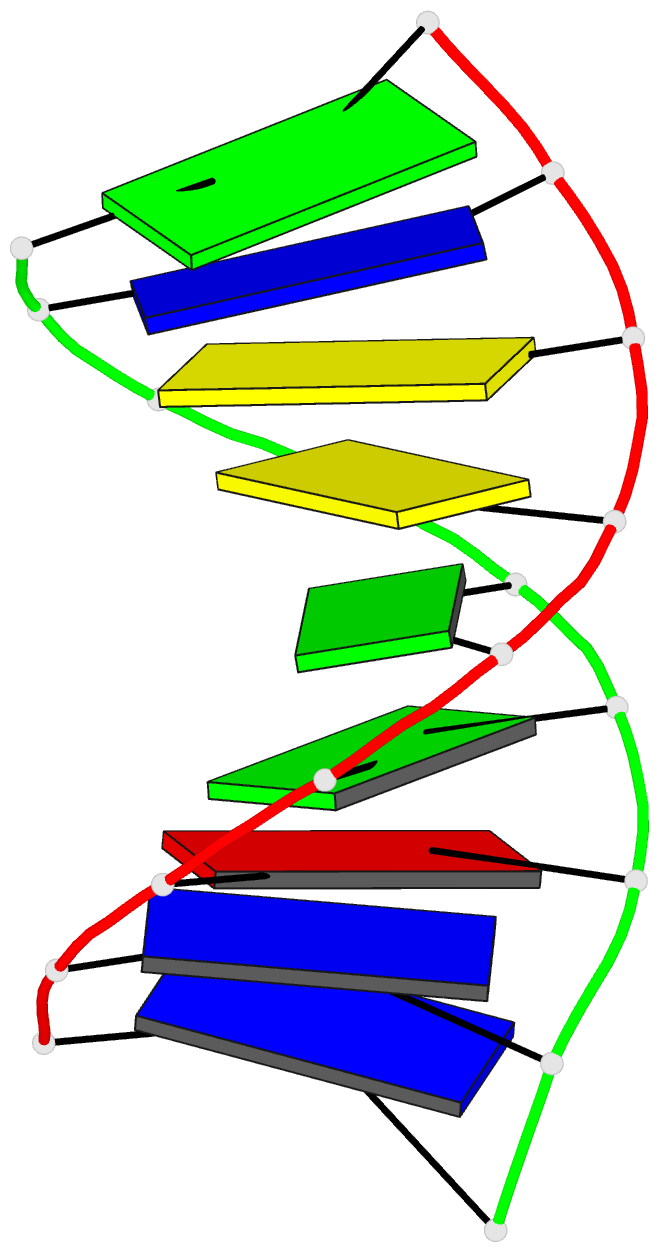
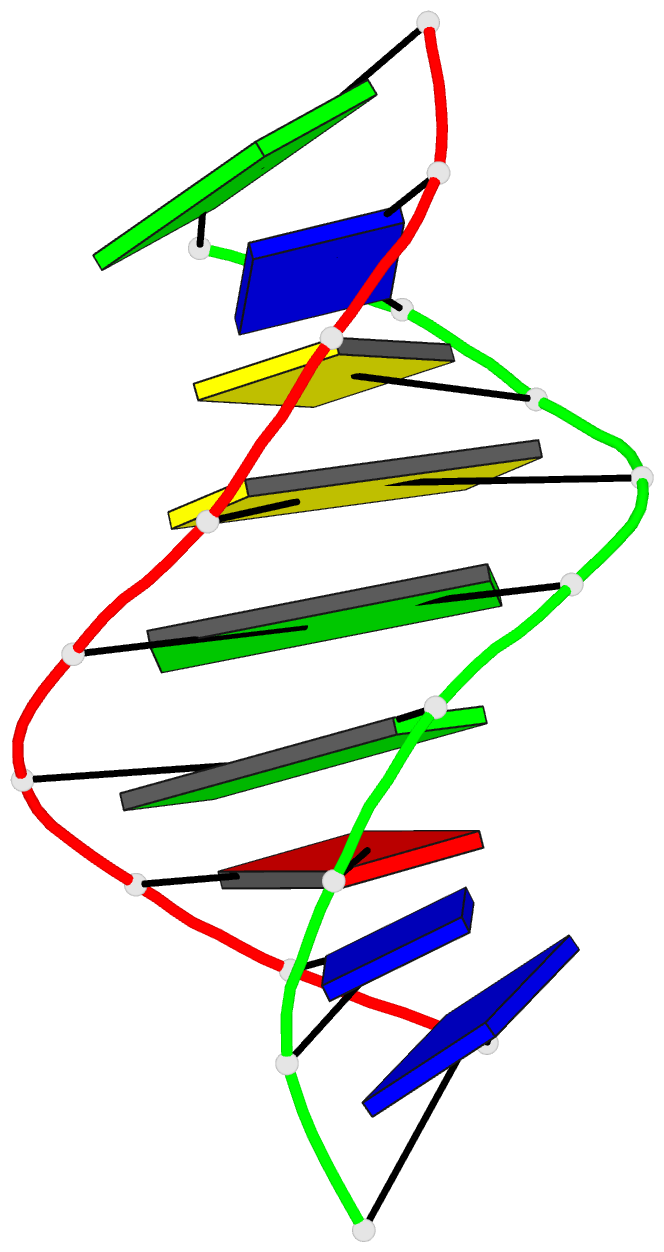
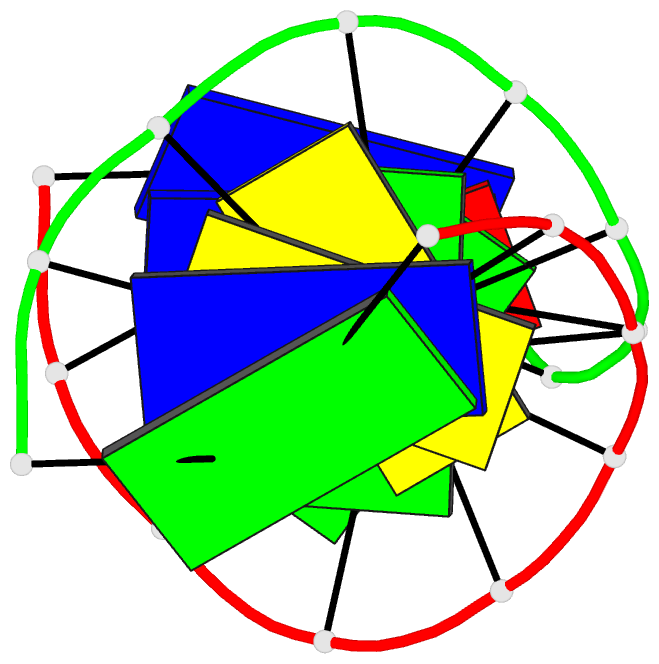
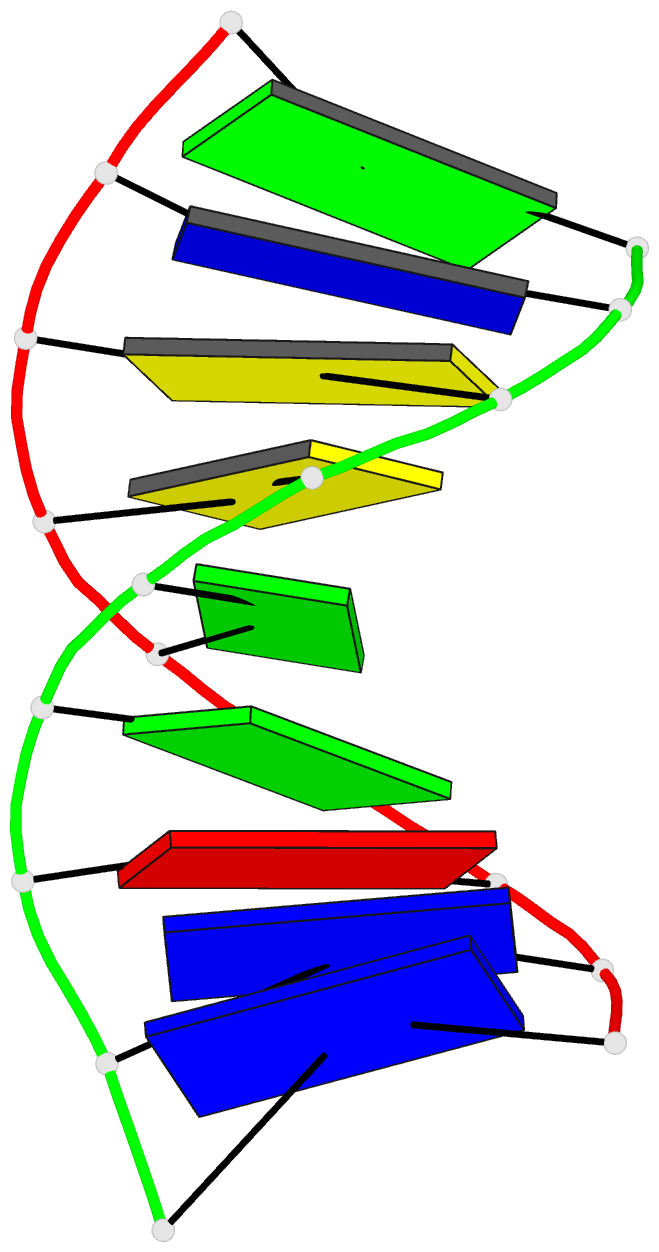
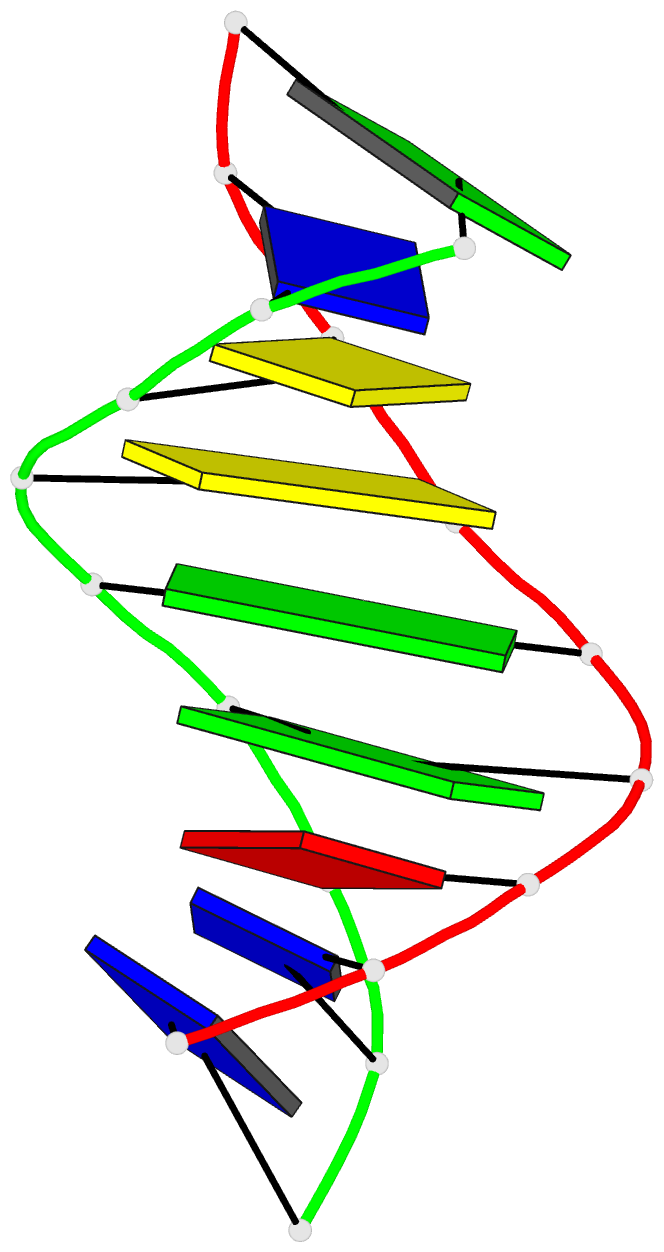
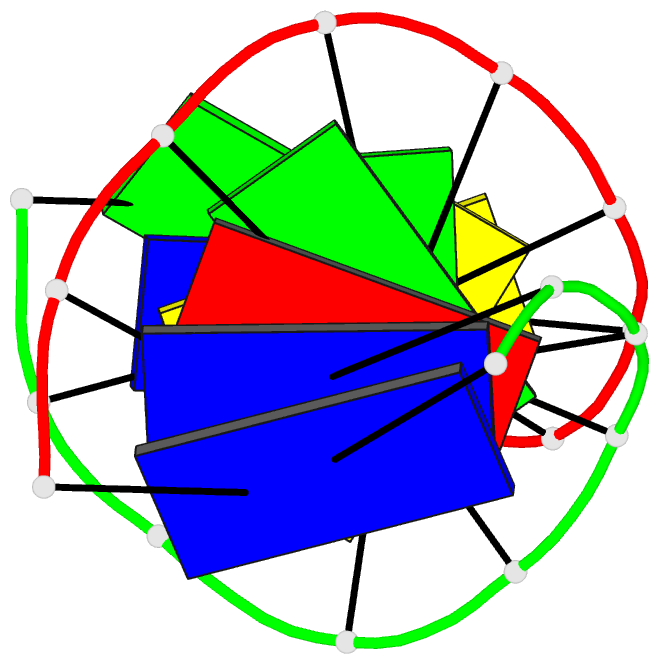
Summary information and primary citation
- PDB-id
- 5ki5; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Structural impact of single ribonucleotides in DNA
- Reference
- Evich M, Spring-Connell AM, Storici F, Germann MW (2016): "Structural Impact of Single Ribonucleotide Residues in DNA." Chembiochem, 17, 1968-1977. doi: 10.1002/cbic.201600385.
- Abstract
- Single ribonucleotide intrusions represent the most common non-standard nucleotide in genomic DNA, yet little is known of their structural impact. This lesion incurs genomic instability in addition to affecting the physical properties of the DNA.To probe for structural and dynamic effects of single ribonucleotides in various sequence contexts, AxC, CxG and GxC, where x=rG or dG, we report the structures of three single ribonucleotide containing DNA duplexes and the corresponding DNA controls. This lesion subtly and locally perturbs the structure asymmetrically 3' of the lesion on both the riboguanosine-containing and complementary strand of the duplex. The perturbations are mainly restricted to the sugar and phosphodiester backbone. The ribose and 3' downstream deoxyriboses are predominately in N-type conformation, while backbone torsion angles epsilon and/or zeta of the ribonucleotide or upstream deoxyribonucleotide are affected. Depending on the flanking sequences, the C2'-OH group forms hydrogen bonds with the backbone, 3'- neighboring base and/or sugar. Interestingly, even in a similar purine-rG-pyrimidine environment (A-rG-C and G-rG-C), a riboguanosine affects DNA in a distinct manner and manifests different hydrogen bonds, which makes generalizations difficult.