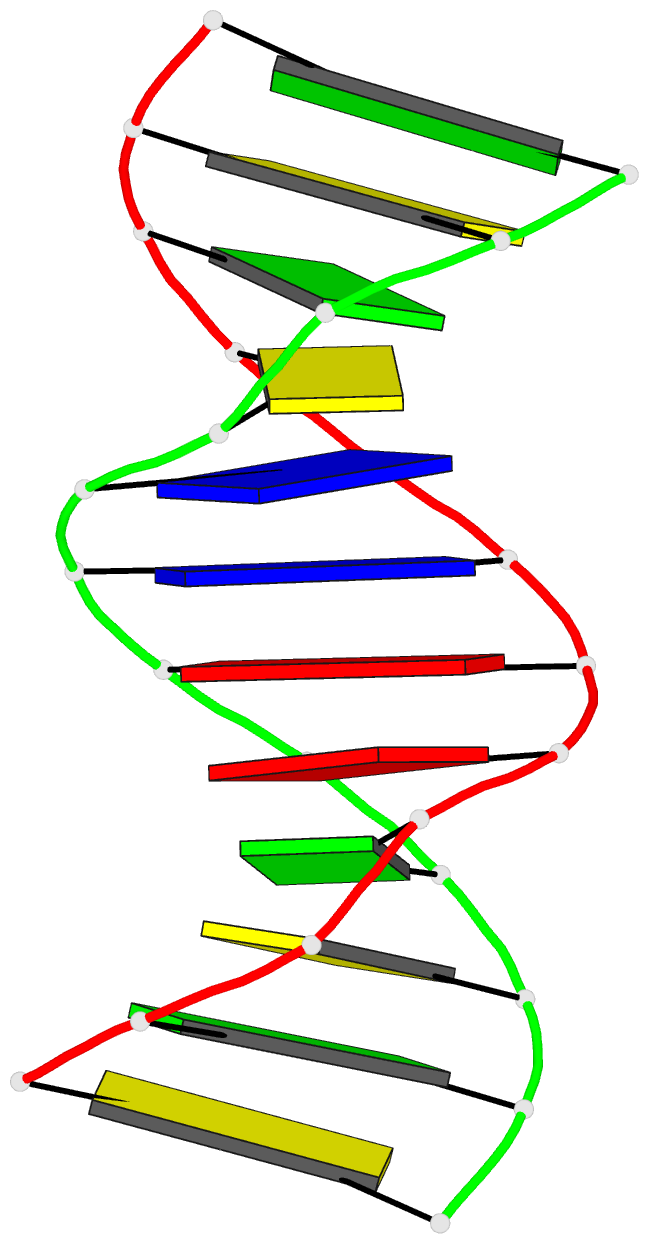
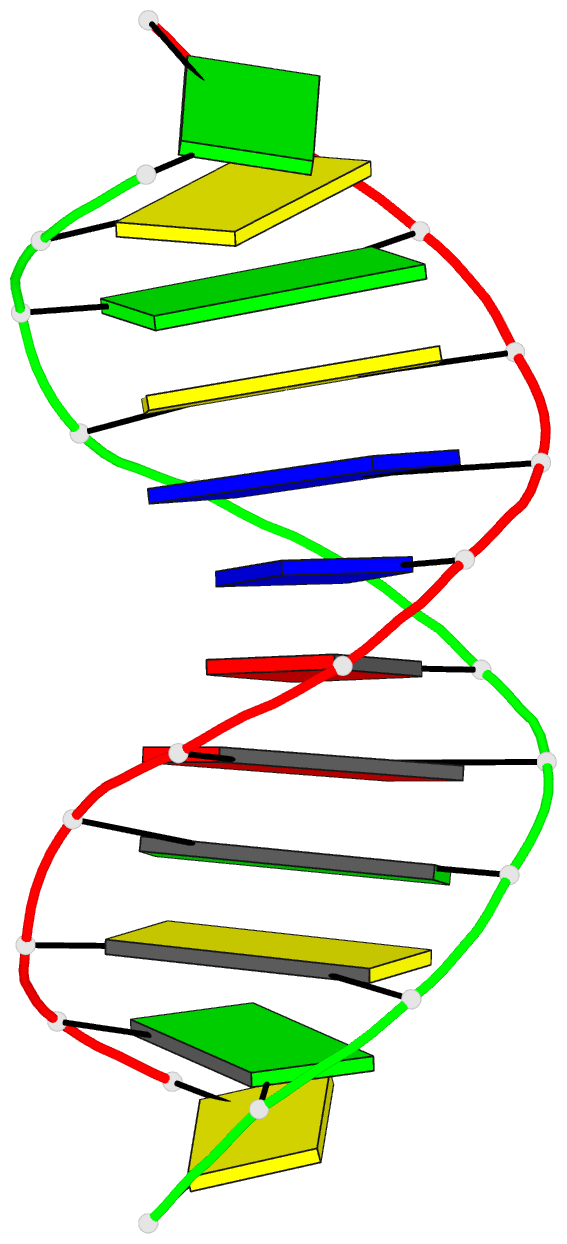
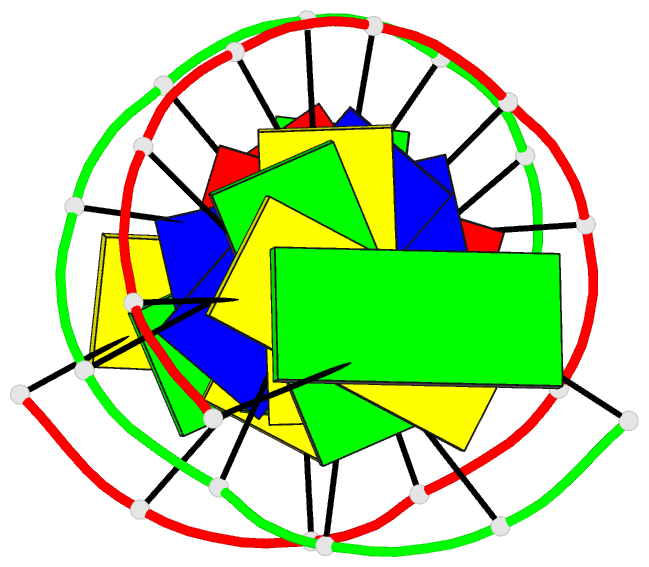
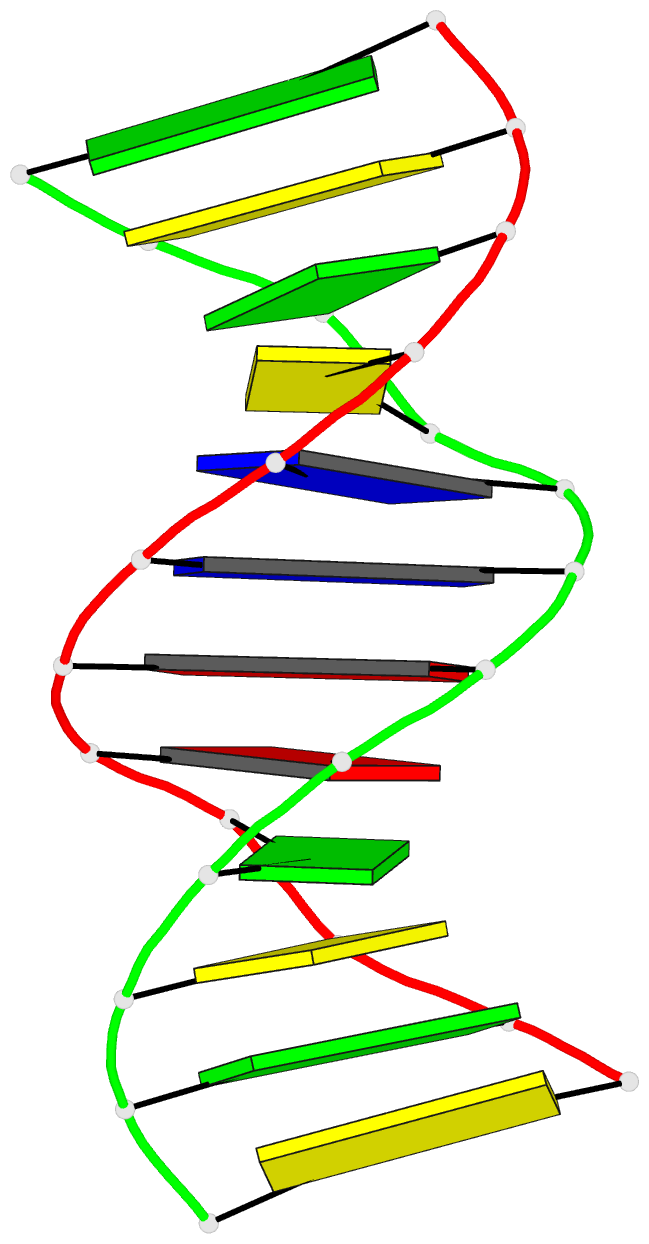
Summary information and primary citation
- PDB-id
- 5iv1; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of DNA dodecamer with 8-oxoguanine at 4th position
- Reference
- Hoppins JJ, Gruber DR, Miears HL, Kiryutin AS, Kasymov RD, Petrova DV, Endutkin AV, Popov AV, Yurkovskaya AV, Fedechkin SO, Brockerman JA, Zharkov DO, Smirnov SL (2016): "8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme." Plos One, 11, e0164424. doi: 10.1371/journal.pone.0164424.
- Abstract
- 8-oxoguanine is one of the most abundant and impactful oxidative DNA lesions. However, the reasons underlying its effects, especially those not directly explained by the altered base pairing ability, are poorly understood. We report the effect of the lesion on the action of EcoRI, a widely used restriction endonuclease. Introduction of 8-oxoguanine inside, or adjacent to, the GAATTC recognition site embedded within the Drew-Dickerson dodecamer sequence notably reduced the EcoRI activity. Solution NMR revealed that 8-oxoguanine in the DNA duplex causes substantial alterations in the sugar-phosphate backbone conformation, inducing a BI→BII transition. Moreover, molecular dynamics of the complex suggested that 8-oxoguanine, although does not disrupt the sequence-specific contacts formed by the enzyme with DNA, shifts the distribution of BI/BII backbone conformers. Based on our data, we propose that the disruption of enzymatic cleavage can be linked with the altered backbone conformation and dynamics in the free oxidized DNA substrate and, possibly, at the protein-DNA interface.