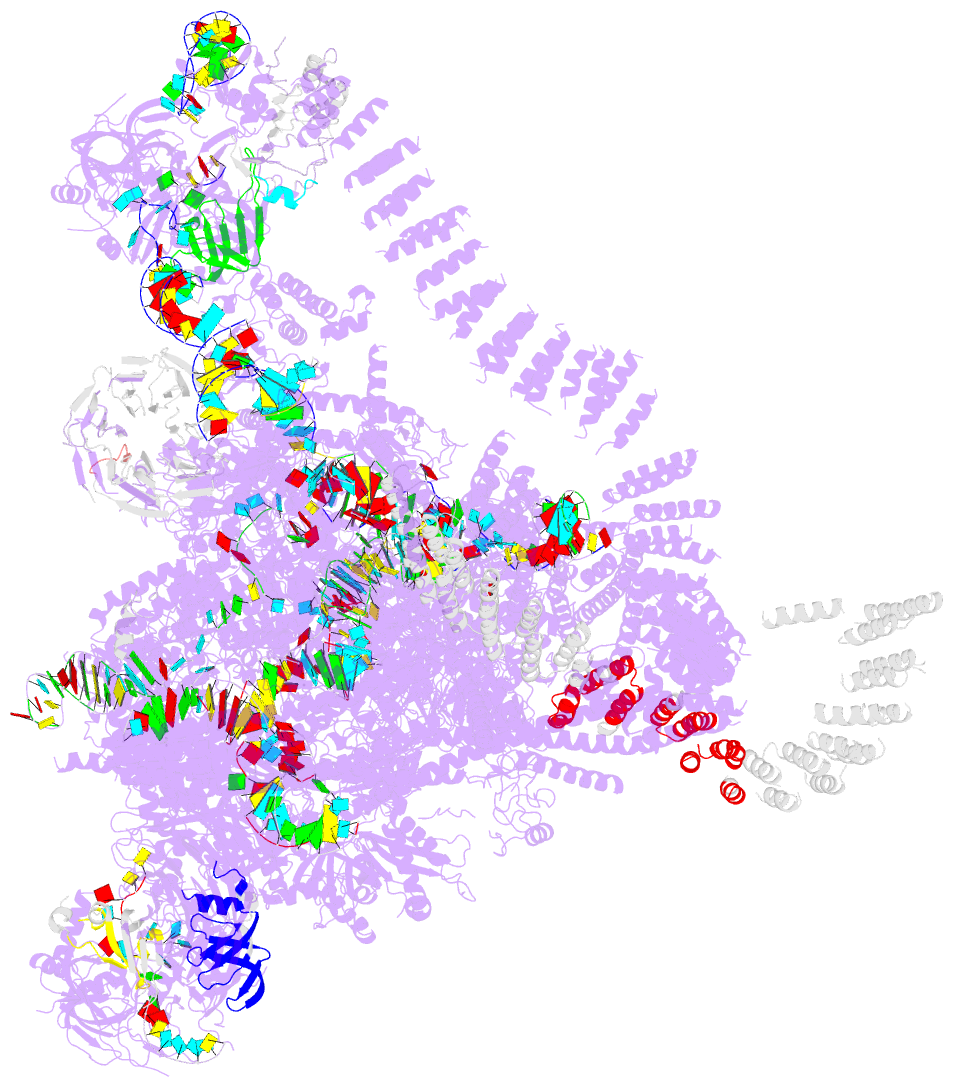
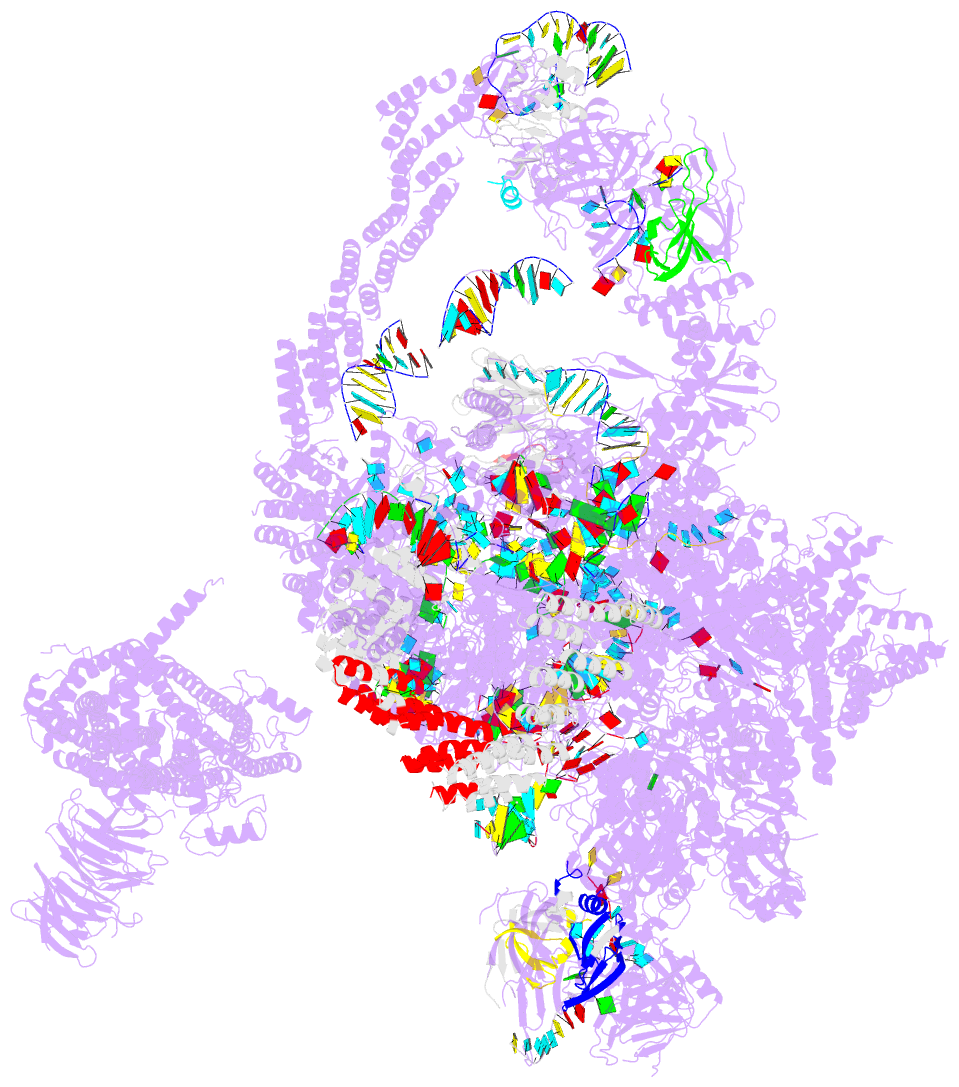
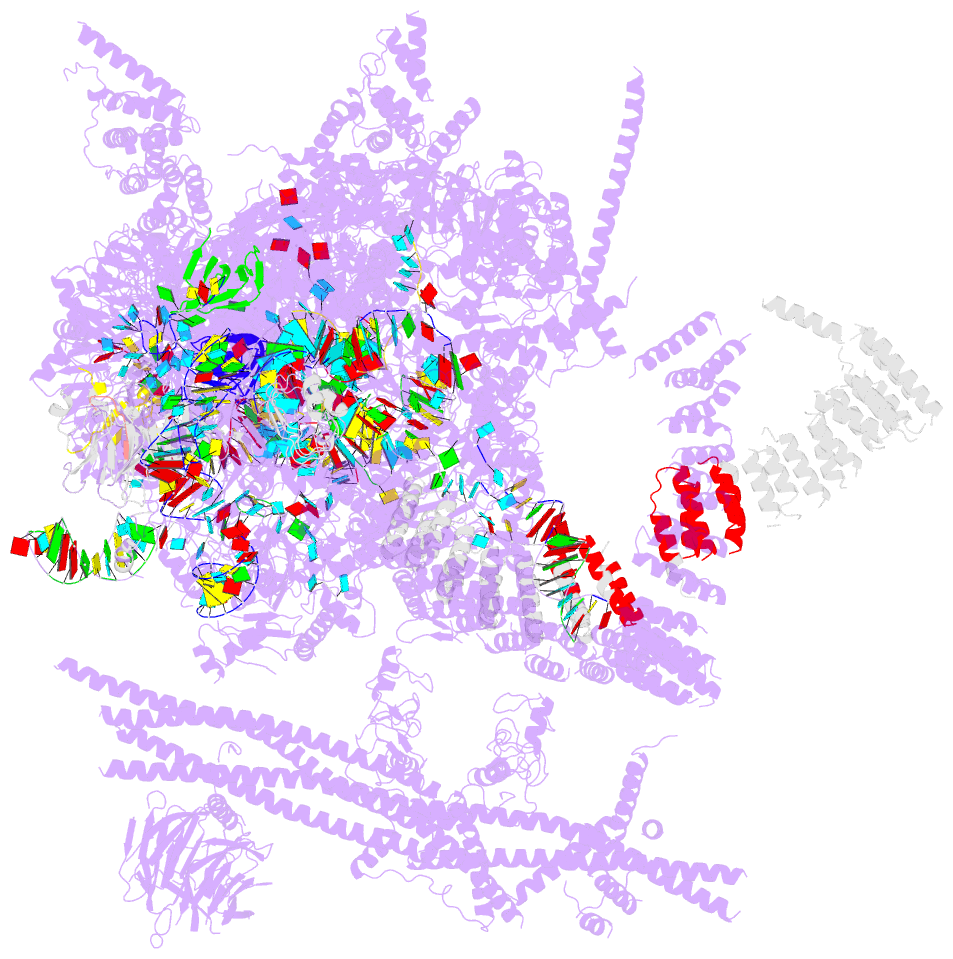
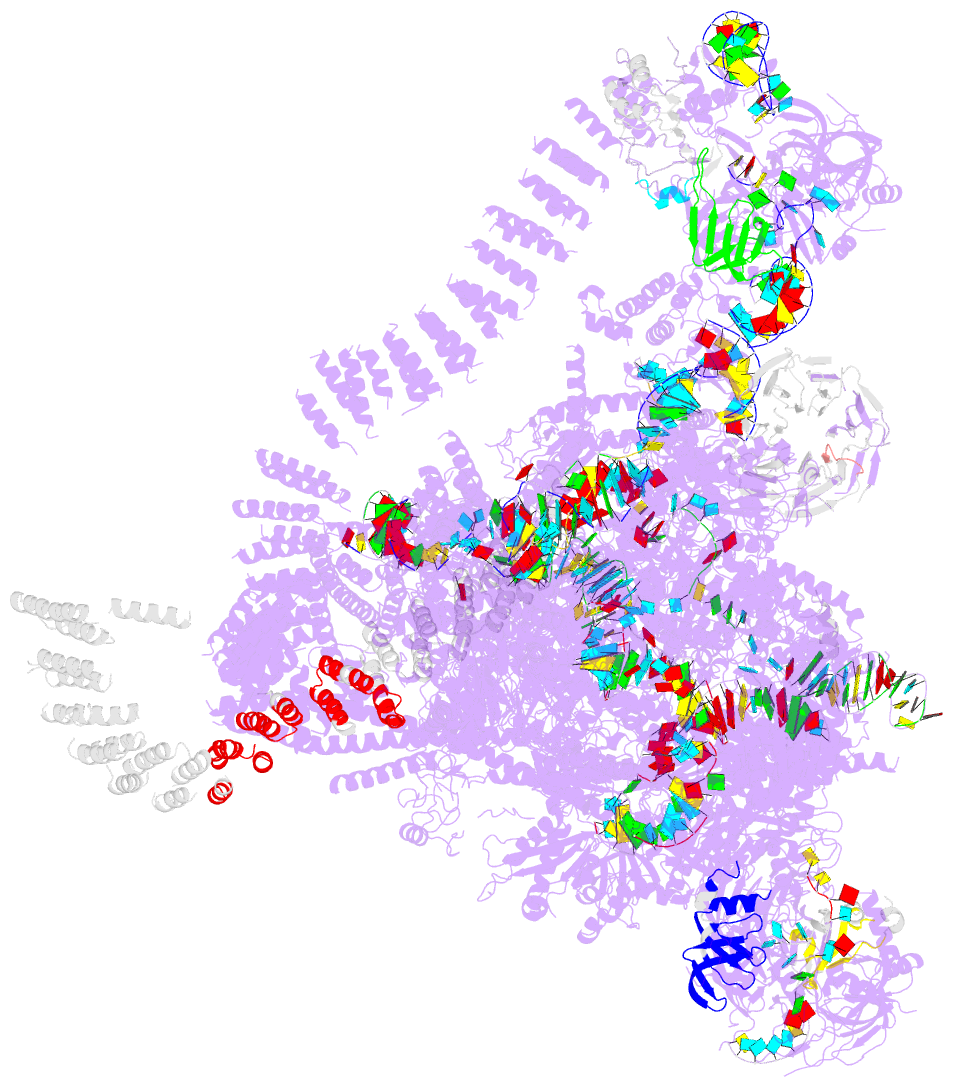
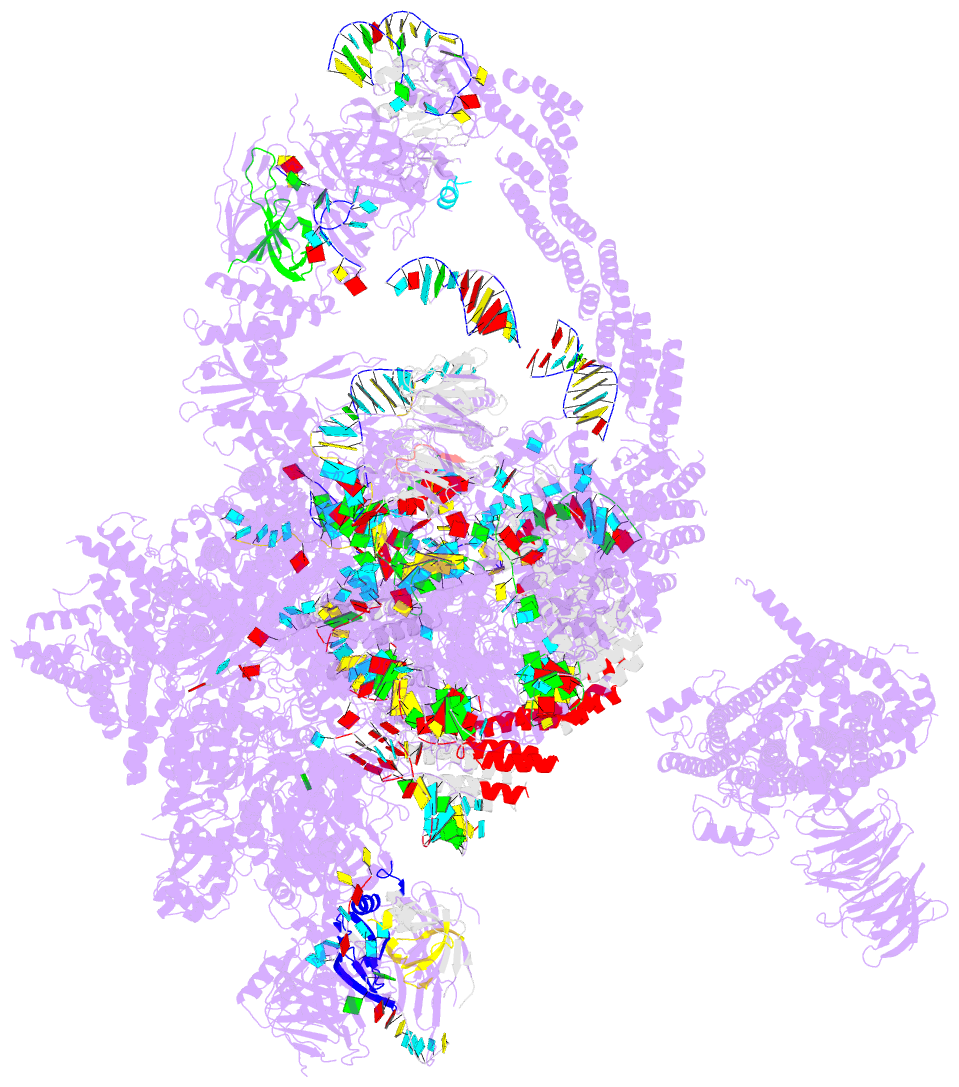
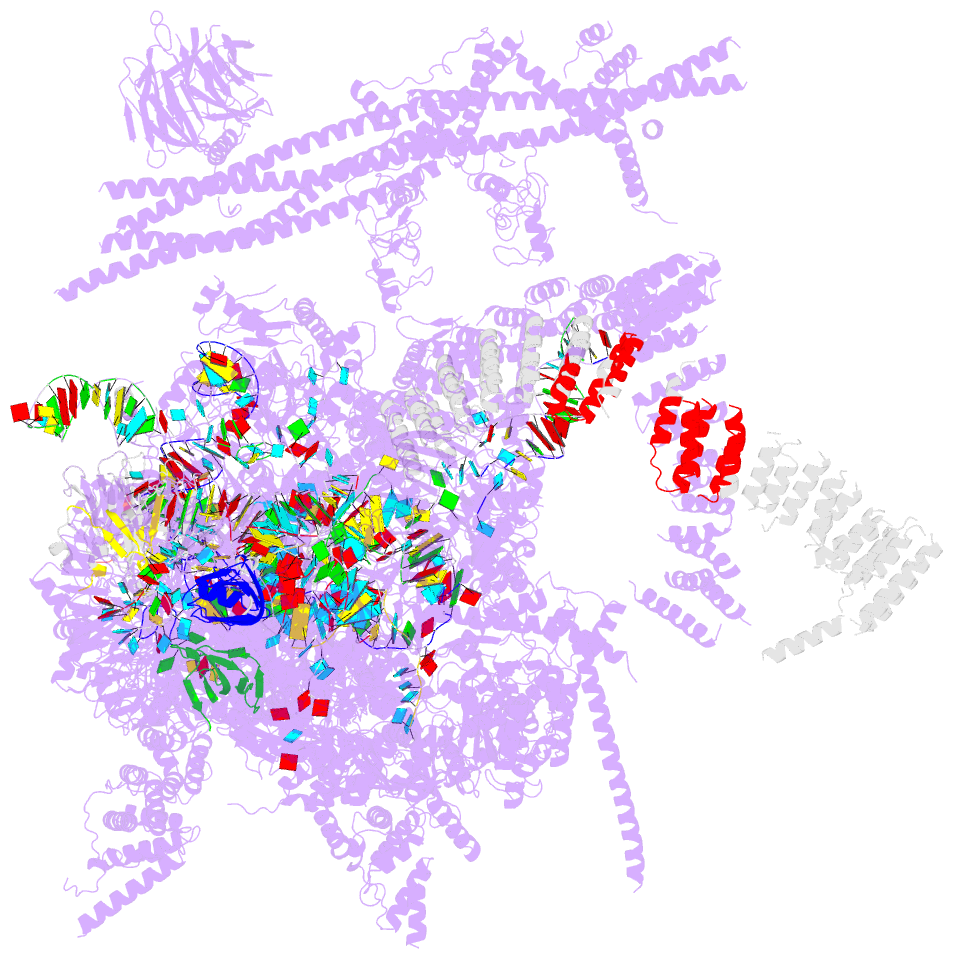
Summary information and primary citation
- PDB-id
- 5gmk; DSSR-derived features in text and JSON formats
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.4 Å)
- Summary
- cryo-EM structure of the catalytic step i spliceosome (c complex) at 3.4 angstrom resolution
- Reference
- Wan R, Yan C, Bai R, Huang G, Shi Y (2016): "Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution." Science, 353, 895-904. doi: 10.1126/science.aag2235.
- Abstract
- Each cycle of pre-mRNA splicing, carried out by the spliceosome, comprises two sequential transesterification reactions, which result in the removal of an intron and joining of two exons. Here we report an atomic structure of a catalytic step I spliceosome (known as the C complex) from Saccharomyces cerevisiae, determined by cryo-electron microscopy at an average resolution of 3.4 Å. In the structure, the 2'-OH of the invariant adenine nucleotide in the branch point sequence (BPS) is covalently joined to the phosphate at the 5'-end of the 5'-splice site (5'SS), forming an intron lariat. The freed 5'-exon remains anchored to loop I of U5 small nuclear RNA (snRNA), and the 5'SS and BPS of the intron form duplexes with conserved U6 and U2 snRNA sequences, respectively. Specific placement of these RNA elements at the catalytic cavity of Prp8 is stabilized by 15 protein components, including Snu114 and the splicing factors Cwc21, Cwc22, Cwc25, and Yju2. These features, representing the conformation of the spliceosome after the first-step reaction, predict structural changes that are needed for the execution of the second-step transesterification reaction.