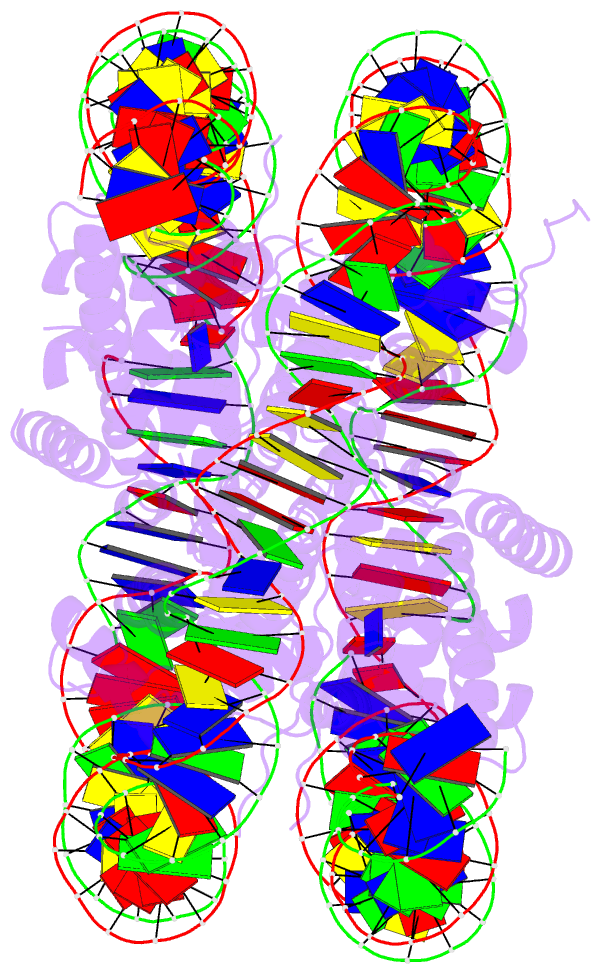
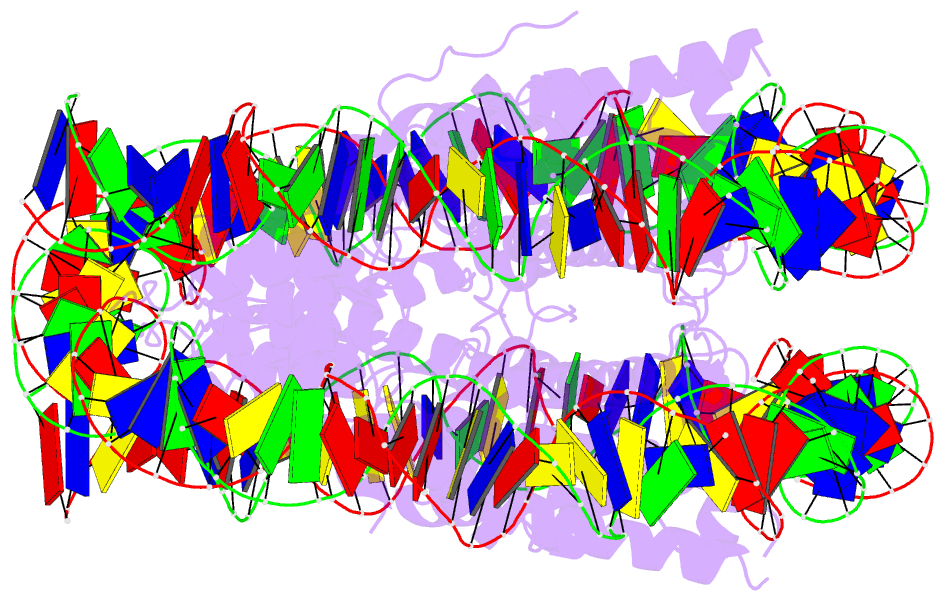
Summary information and primary citation
- PDB-id
- 4ym5; DSSR-derived features in text and JSON formats
- Class
- structural protein-DNA
- Method
- X-ray (4.005 Å)
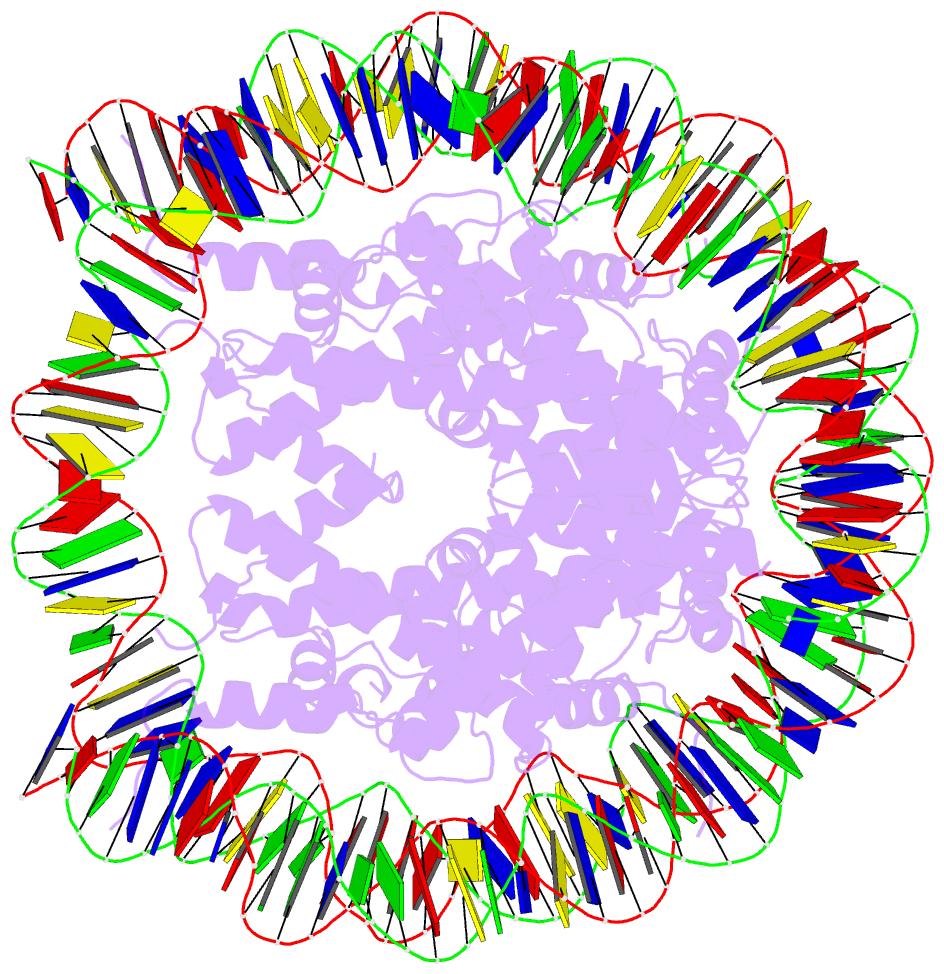
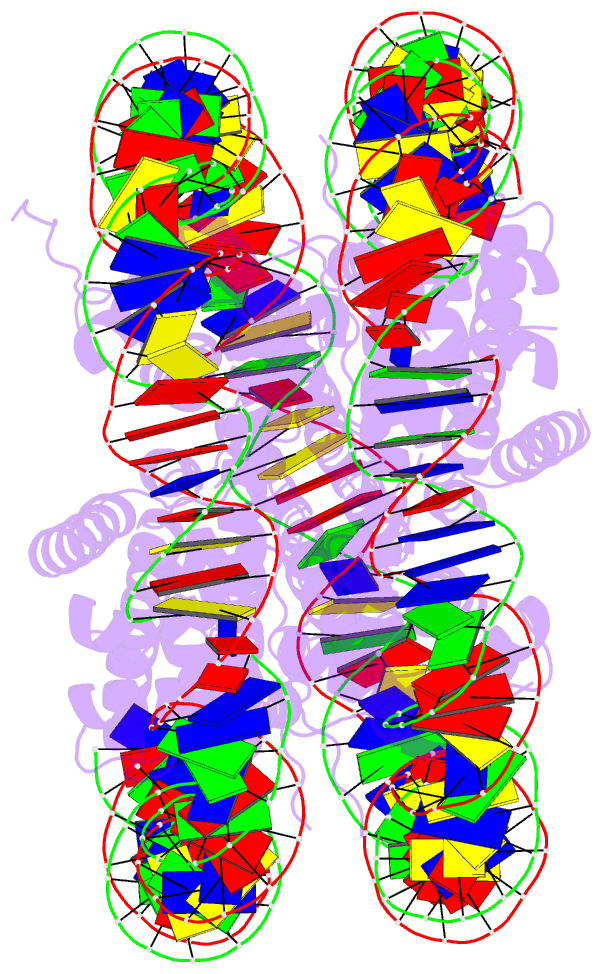
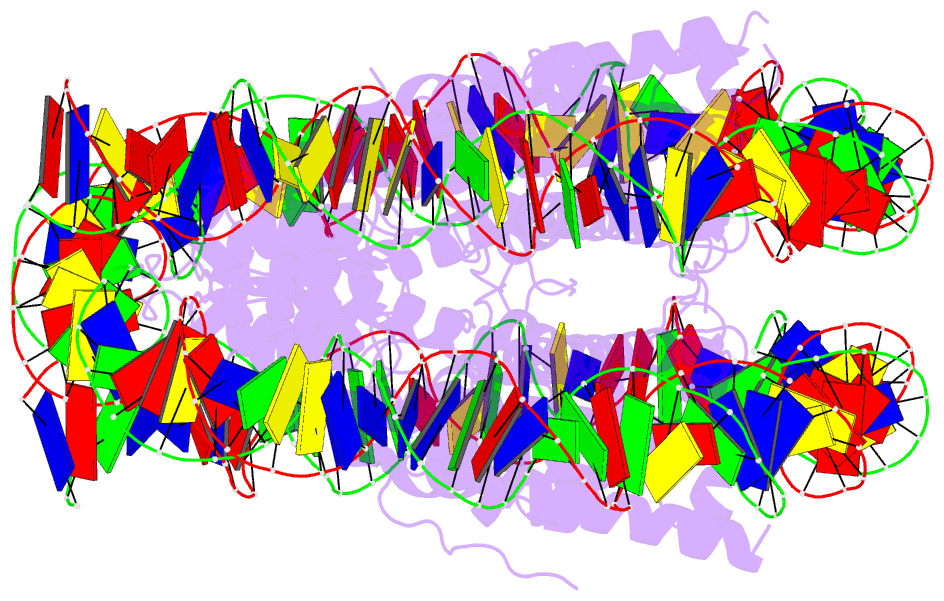
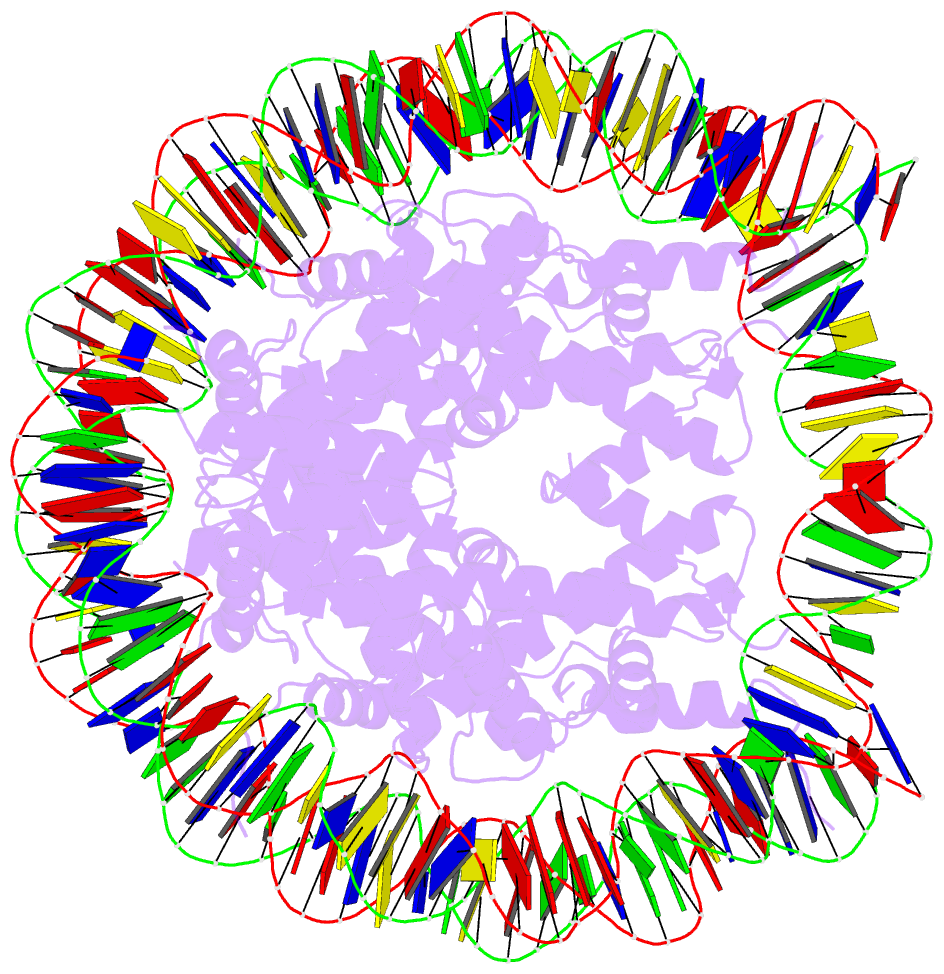
- Summary
- Crystal structure of the human nucleosome containing 6-4pp (inside)
- Reference
- Osakabe A, Tachiwana H, Kagawa W, Horikoshi N, Matsumoto S, Hasegawa M, Matsumoto N, Toga T, Yamamoto J, Hanaoka F, Thoma NH, Sugasawa K, Iwai S, Kurumizaka H (2015): "Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome." Sci Rep, 5, 16330. doi: 10.1038/srep16330.
- Abstract
- UV-DDB, an initiation factor for the nucleotide excision repair pathway, recognizes 6-4PP lesions through a base flipping mechanism. As genomic DNA is almost entirely accommodated within nucleosomes, the flipping of the 6-4PP bases is supposed to be extremely difficult if the lesion occurs in a nucleosome, especially on the strand directly contacting the histone surface. Here we report that UV-DDB binds efficiently to nucleosomal 6-4PPs that are rotationally positioned on the solvent accessible or occluded surface. We determined the crystal structures of nucleosomes containing 6-4PPs in these rotational positions, and found that the 6-4PP DNA regions were flexibly disordered, especially in the strand exposed to the solvent. This characteristic of 6-4PP may facilitate UV-DDB binding to the damaged nucleosome. We present the first atomic-resolution pictures of the detrimental DNA cross-links of neighboring pyrimidine bases within the nucleosome, and provide the mechanistic framework for lesion recognition by UV-DDB in chromatin.