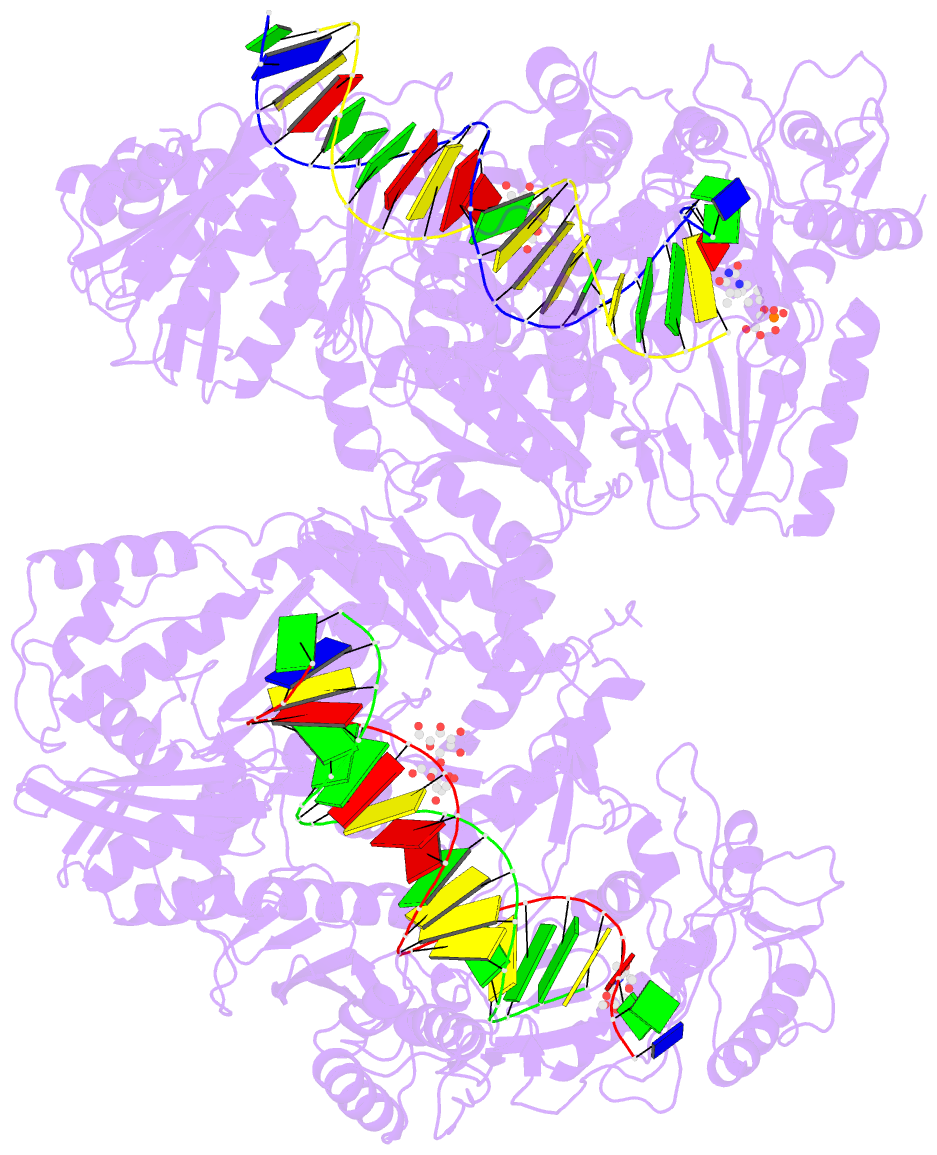
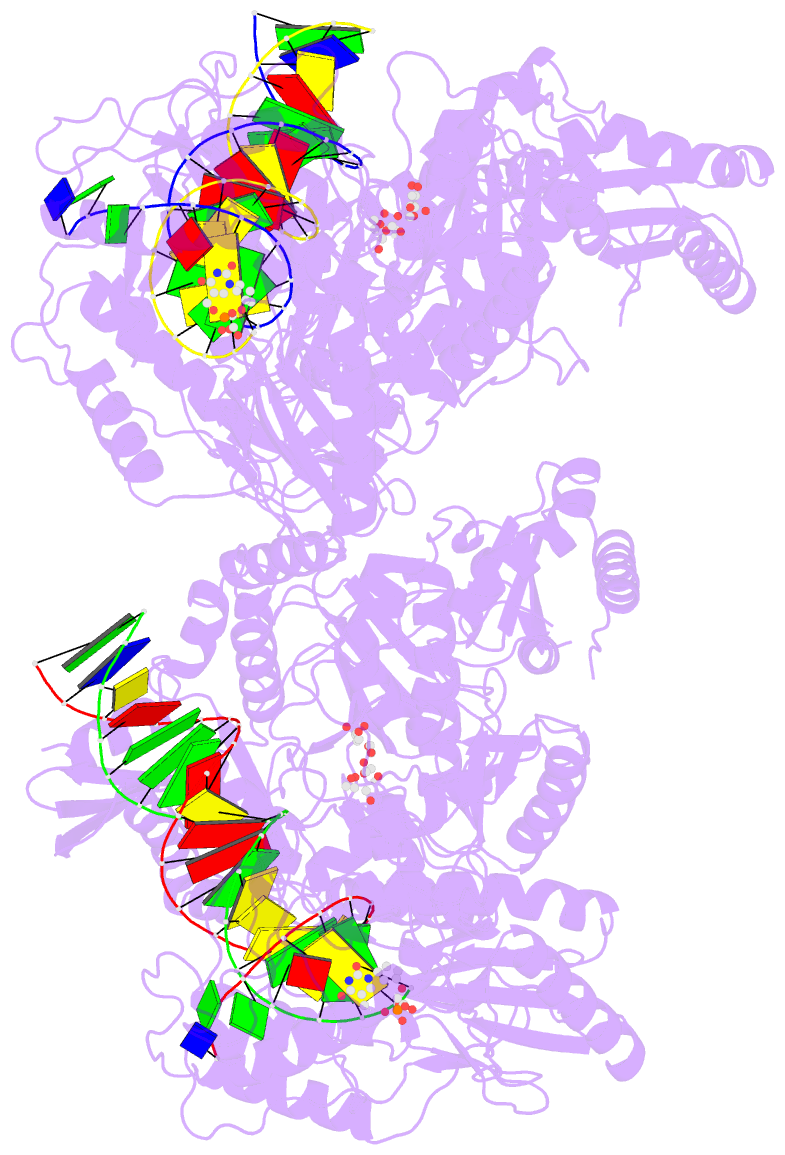
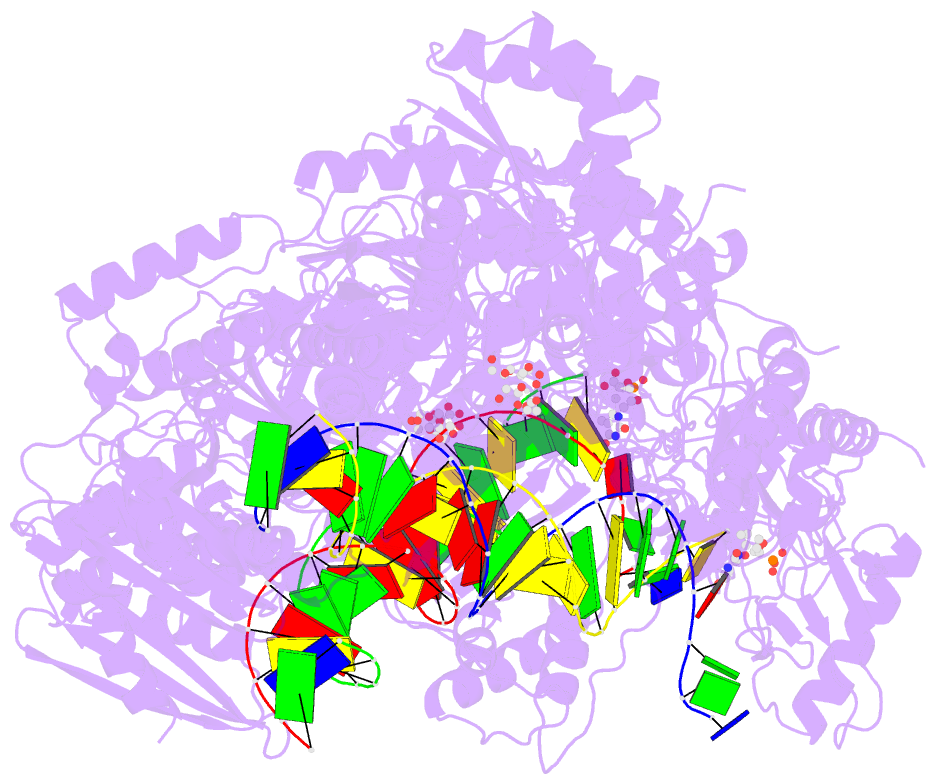
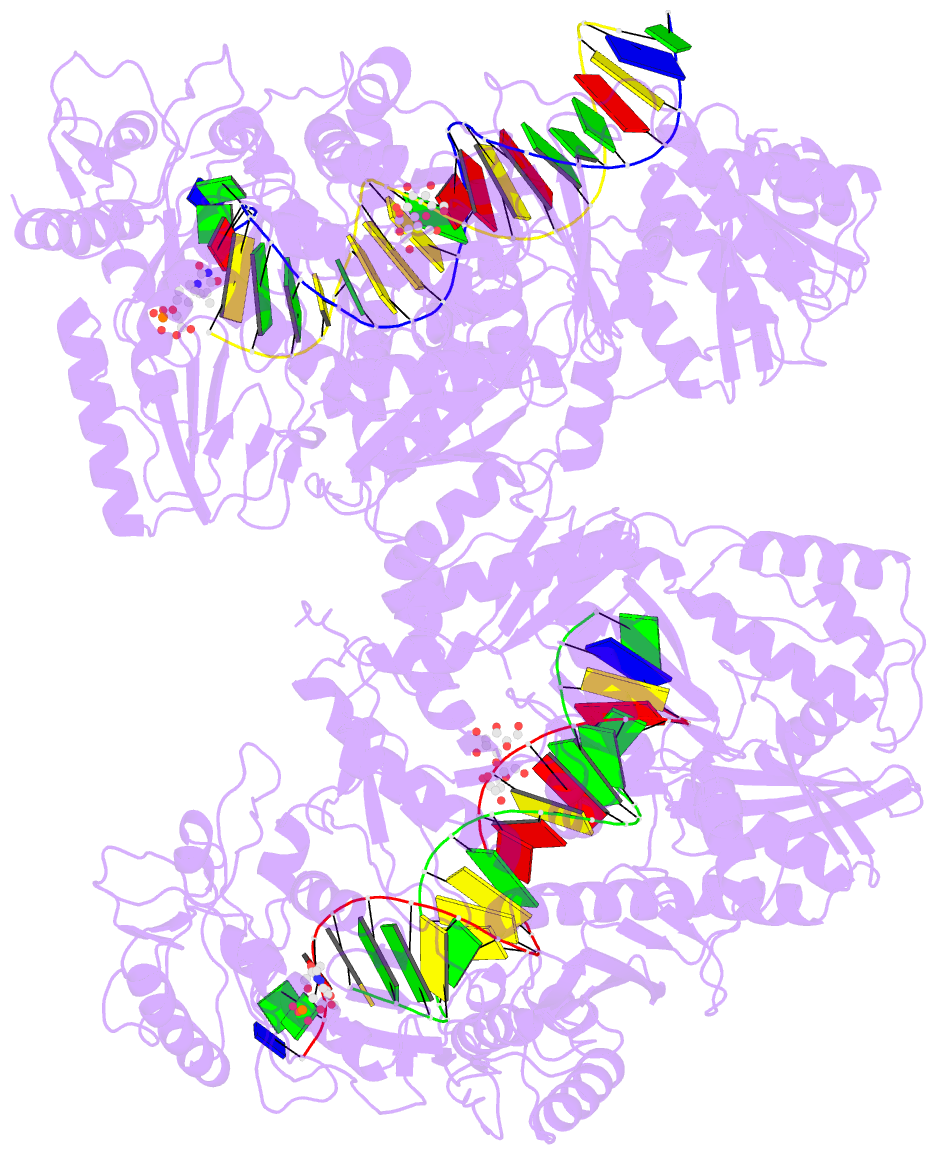
Summary information and primary citation
- PDB-id
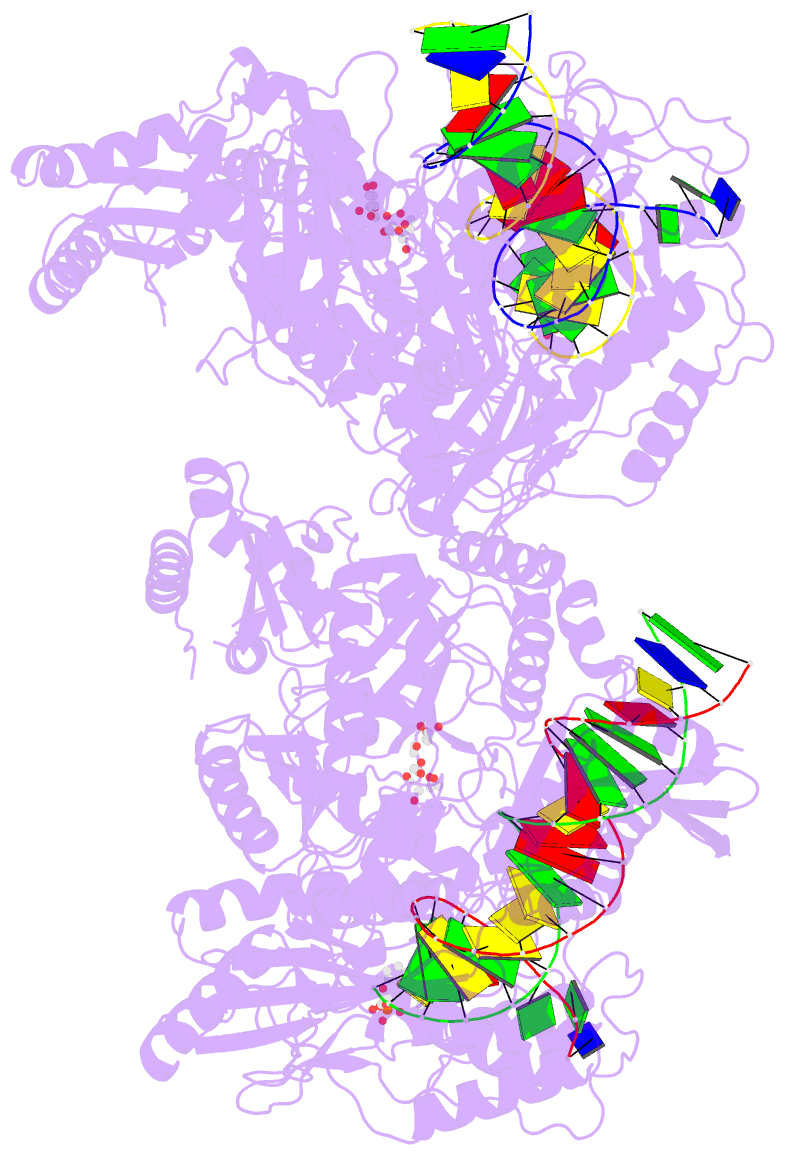
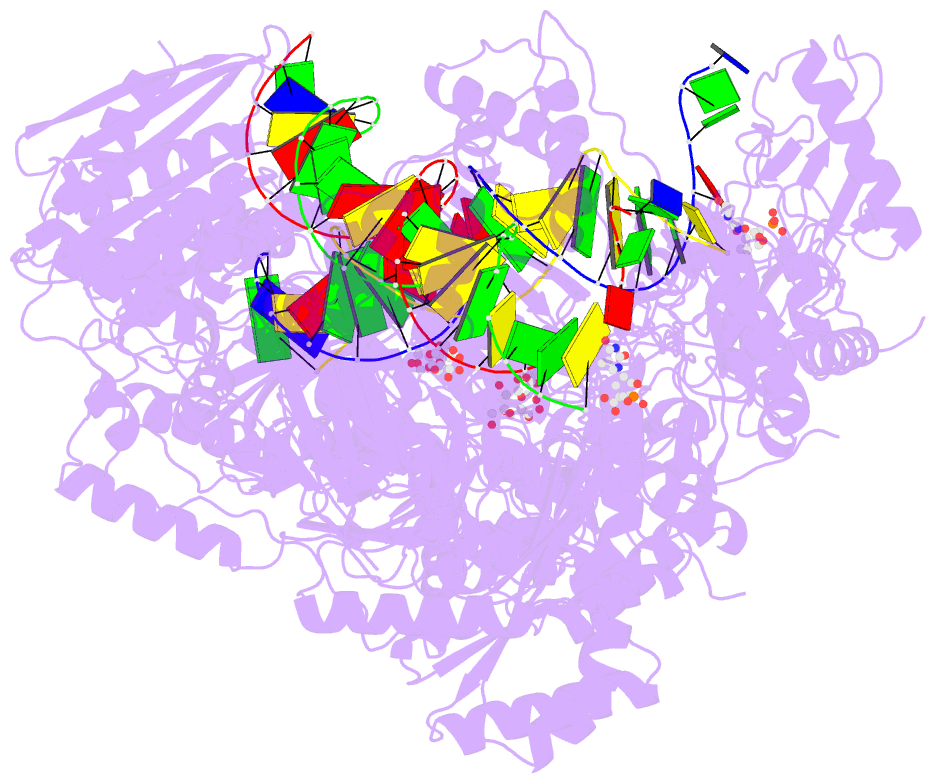
- 4r5p; DSSR-derived features in text and JSON formats
- Class
- transferase, hydrolase-DNA-inhibitor
- Method
- X-ray (2.894 Å)
- Summary
- Crystal structure of hiv-1 reverse transcriptase (rt) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor
- Reference
- Balzarini J, Das K, Bernatchez JA, Martinez SE, Ngure M, Keane S, Ford A, Maguire N, Mullins N, John J, Kim Y, Dehaen W, Vande Voorde J, Liekens S, Naesens L, Gotte M, Maguire AR, Arnold E (2015): "Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics." Proc.Natl.Acad.Sci.USA, 112, 3475-3480. doi: 10.1073/pnas.1420233112.
- Abstract
- Polymerases have a structurally highly conserved negatively charged amino acid motif that is strictly required for Mg(2+) cation-dependent catalytic incorporation of (d)NTP nucleotides into nucleic acids. Based on these characteristics, a nucleoside monophosphonate scaffold, α-carboxy nucleoside phosphonate (α-CNP), was designed that is recognized by a variety of polymerases. Kinetic, biochemical, and crystallographic studies with HIV-1 reverse transcriptase revealed that α-CNPs mimic the dNTP binding through a carboxylate oxygen, two phosphonate oxygens, and base-pairing with the template. In particular, the carboxyl oxygen of the α-CNP acts as the potential equivalent of the α-phosphate oxygen of dNTPs and two oxygens of the phosphonate group of the α-CNP chelate Mg(2+), mimicking the chelation by the β- and γ-phosphate oxygens of dNTPs. α-CNPs (i) do not require metabolic activation (phosphorylation), (ii) bind directly to the substrate-binding site, (iii) chelate one of the two active site Mg(2+) ions, and (iv) reversibly inhibit the polymerase catalytic activity without being incorporated into nucleic acids. In addition, α-CNPs were also found to selectively interact with regulatory (i.e., allosteric) Mg(2+)-dNTP-binding sites of nucleos(t)ide-metabolizing enzymes susceptible to metabolic regulation. α-CNPs represent an entirely novel and broad technological platform for the development of specific substrate active- or regulatory-site inhibitors with therapeutic potential.