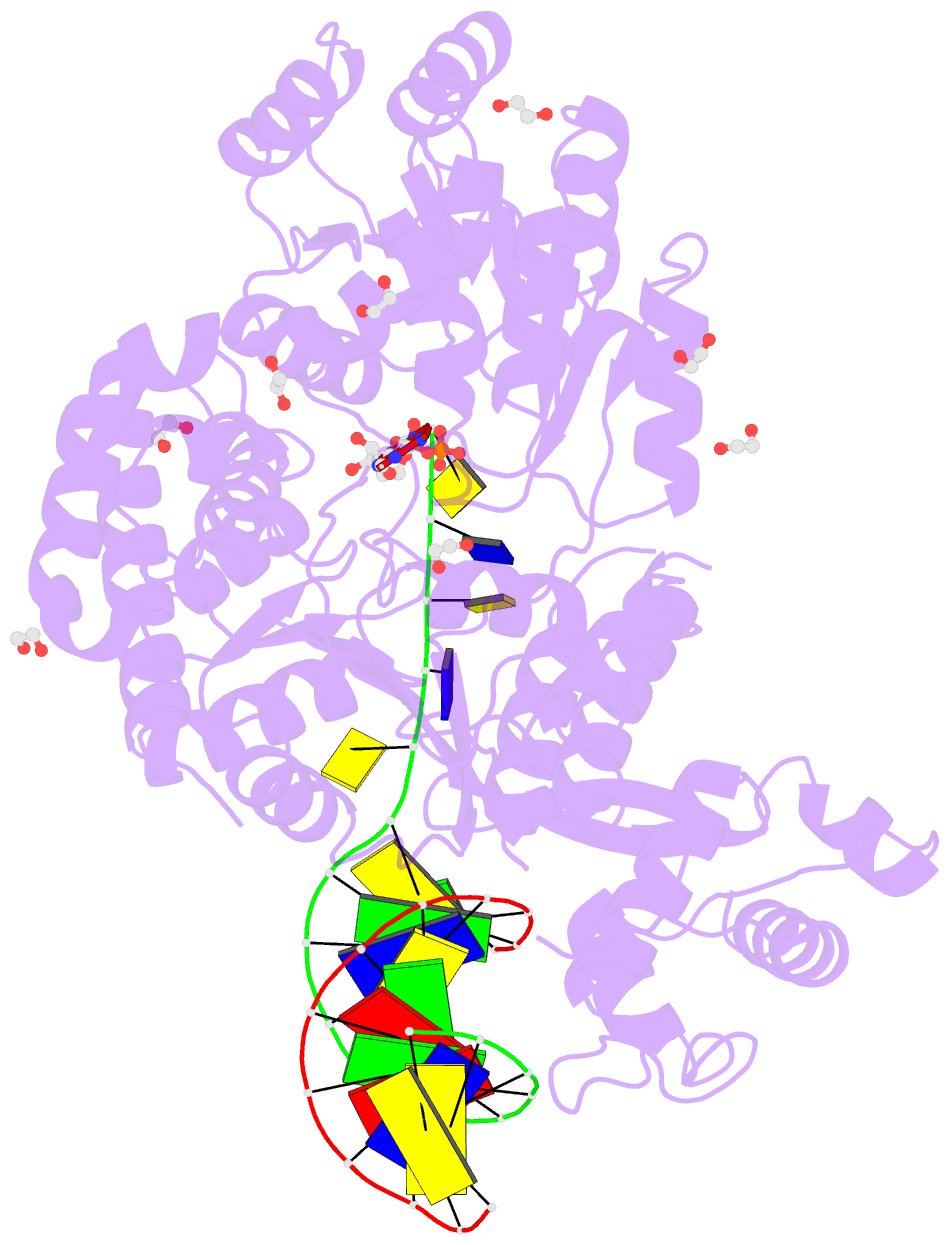
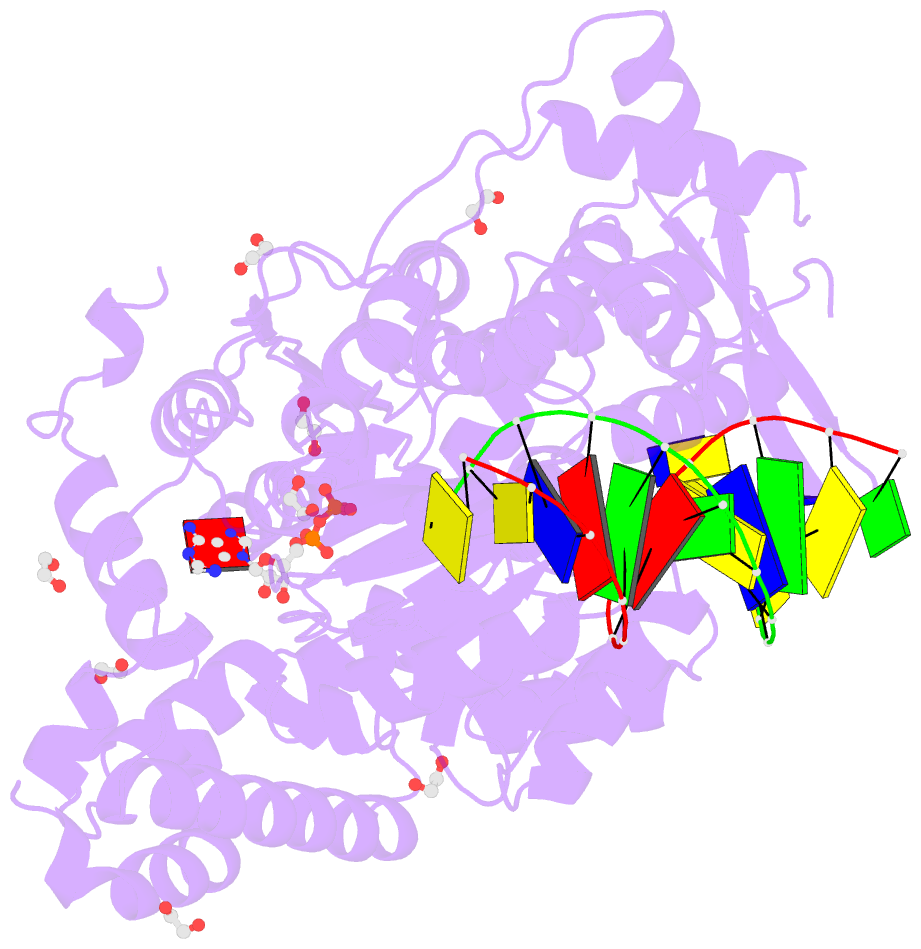
Summary information and primary citation
- PDB-id
- 4o3m; DSSR-derived features in text and JSON formats
- Class
- hydrolase-DNA
- Method
- X-ray (2.3 Å)
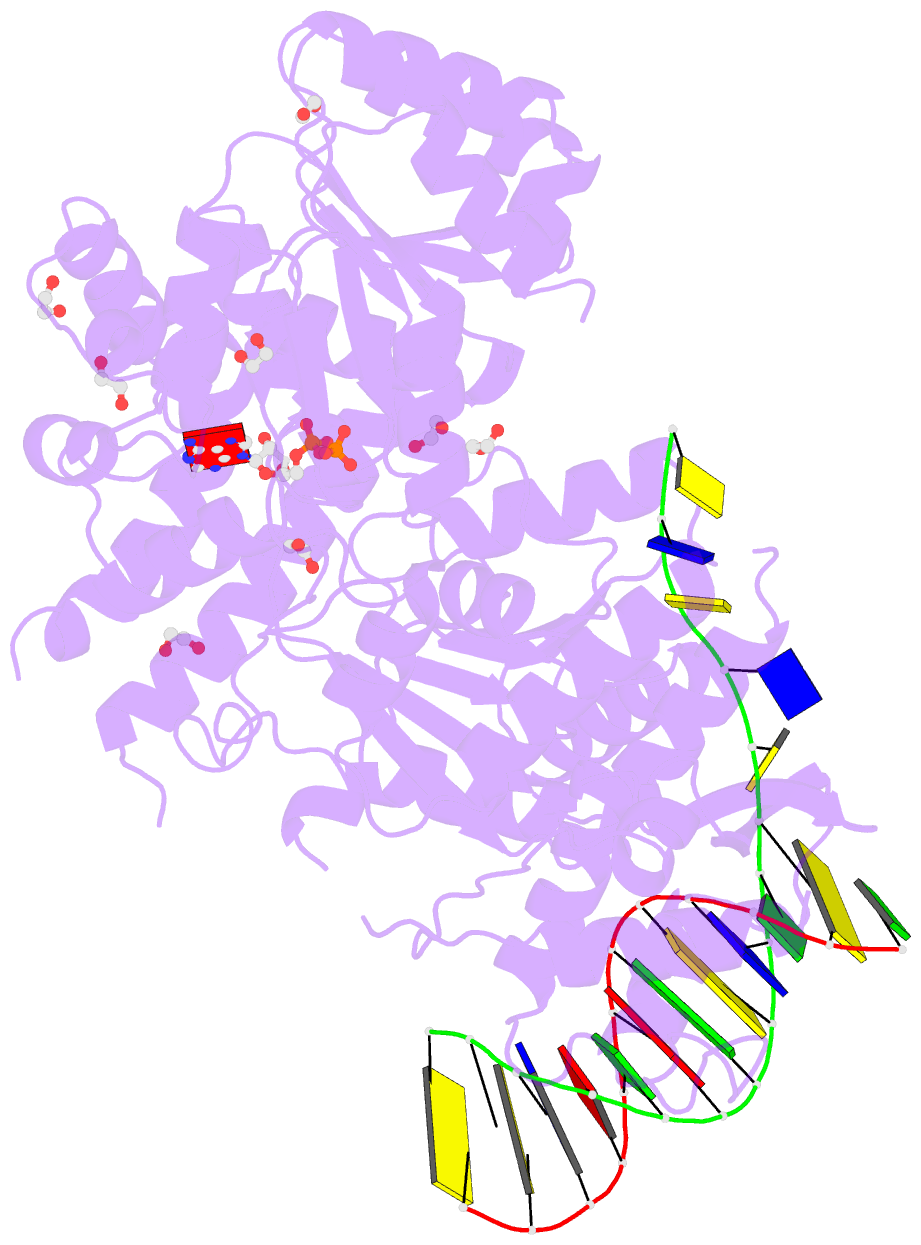
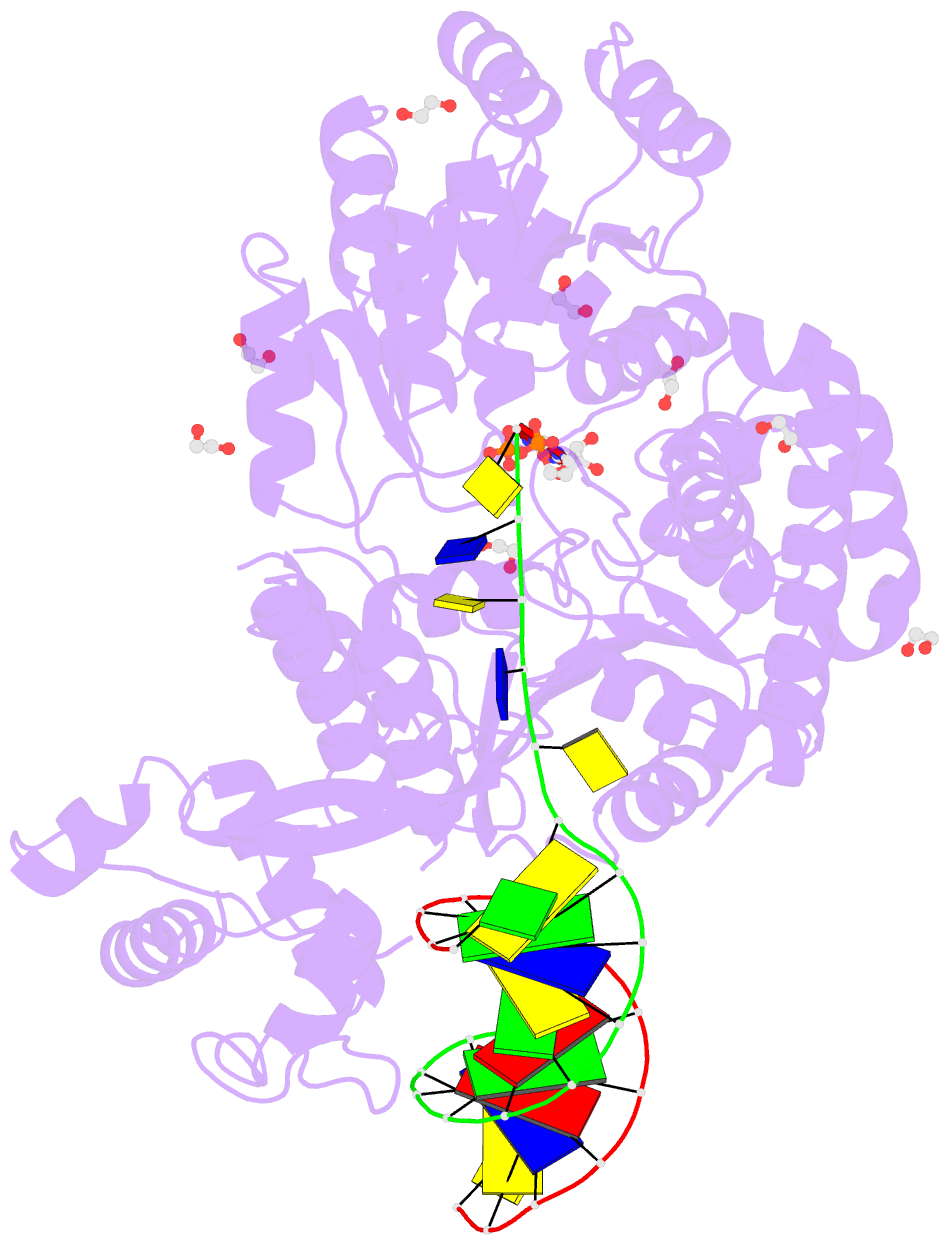
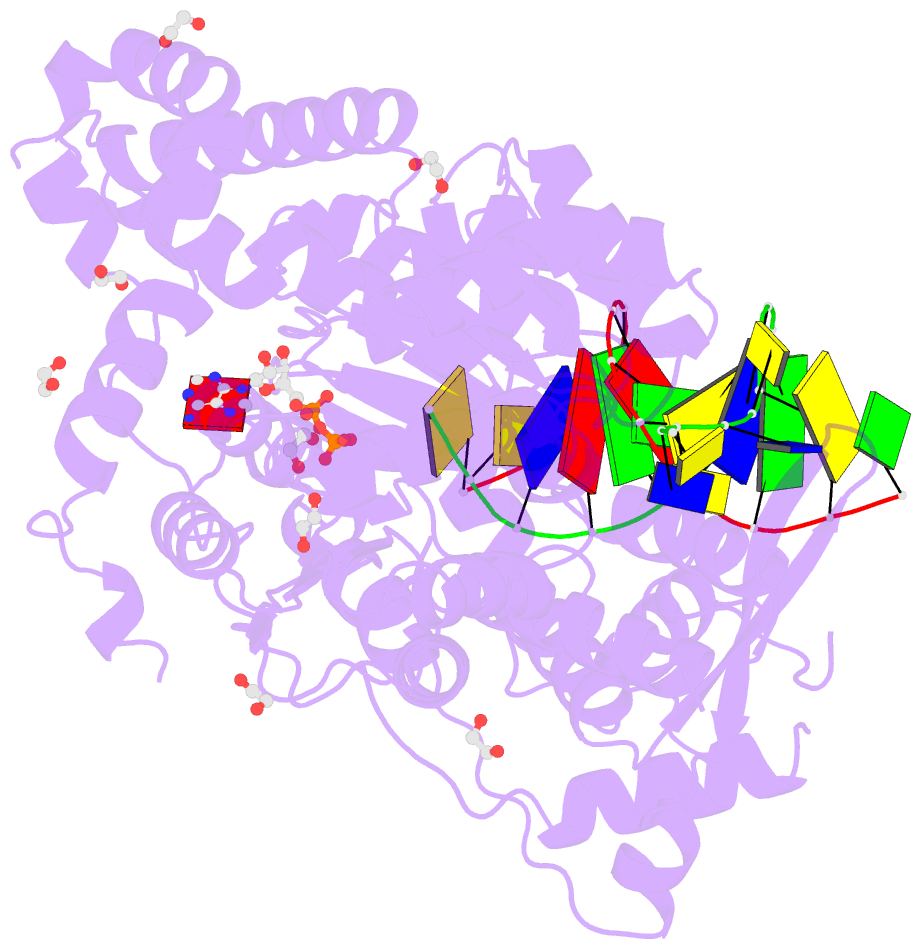
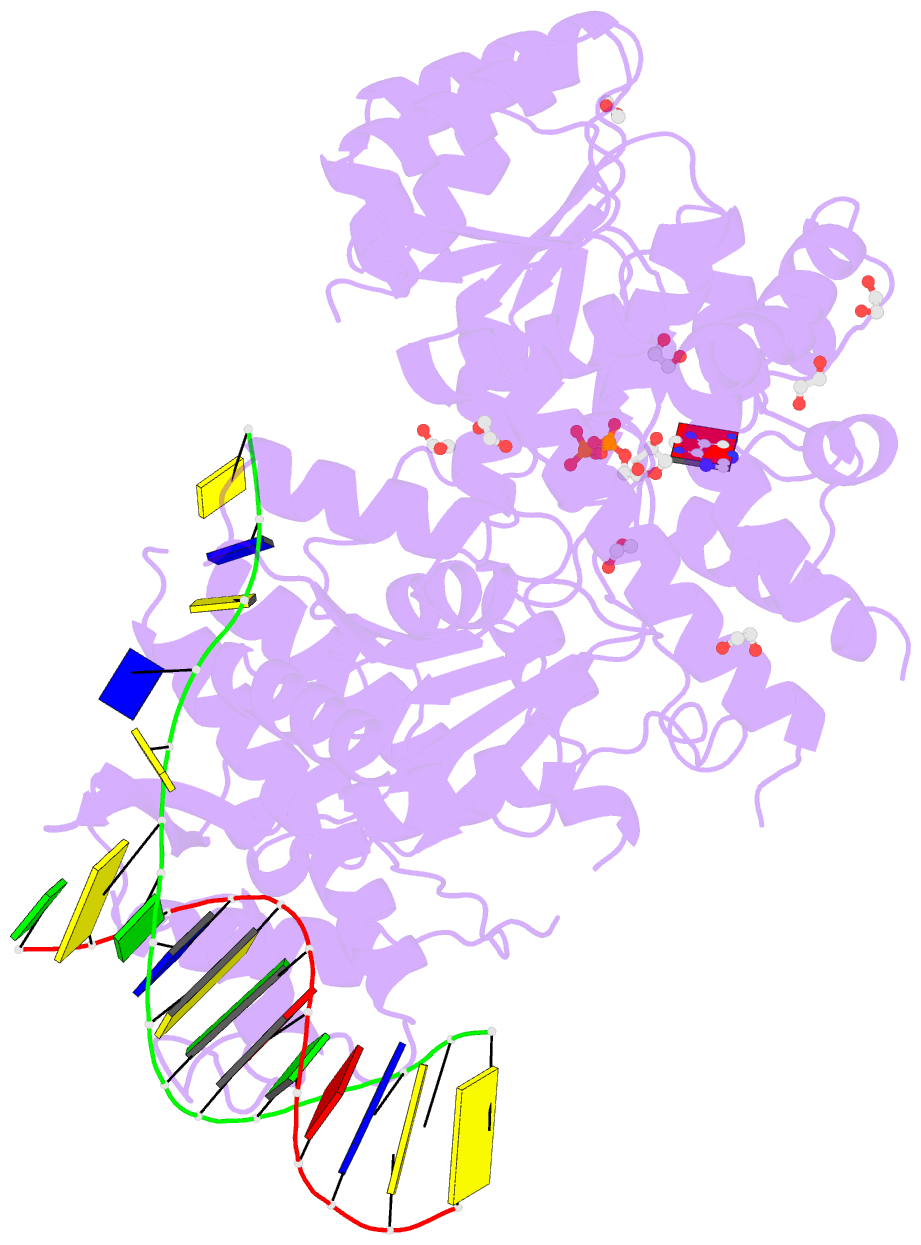
- Summary
- Ternary complex of bloom's syndrome helicase
- Reference
- Swan MK, Legris V, Tanner A, Reaper PM, Vial S, Bordas R, Pollard JR, Charlton PA, Golec JM, Bertrand JA (2014): "Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA." Acta Crystallogr.,Sect.D, 70, 1465-1475. doi: 10.1107/S139900471400501X.
- Abstract
- Bloom's syndrome is an autosomal recessive genome-instability disorder associated with a predisposition to cancer, premature aging and developmental abnormalities. It is caused by mutations that inactivate the DNA helicase activity of the BLM protein or nullify protein expression. The BLM helicase has been implicated in the alternative lengthening of telomeres (ALT) pathway, which is essential for the limitless replication of some cancer cells. This pathway is used by 10-15% of cancers, where inhibitors of BLM are expected to facilitate telomere shortening, leading to apoptosis or senescence. Here, the crystal structure of the human BLM helicase in complex with ADP and a 3'-overhang DNA duplex is reported. In addition to the helicase core, the BLM construct used for crystallization (residues 640-1298) includes the RecQ C-terminal (RQC) and the helicase and ribonuclease D C-terminal (HRDC) domains. Analysis of the structure provides detailed information on the interactions of the protein with DNA and helps to explain the mechanism coupling ATP hydrolysis and DNA unwinding. In addition, mapping of the missense mutations onto the structure provides insights into the molecular basis of Bloom's syndrome.