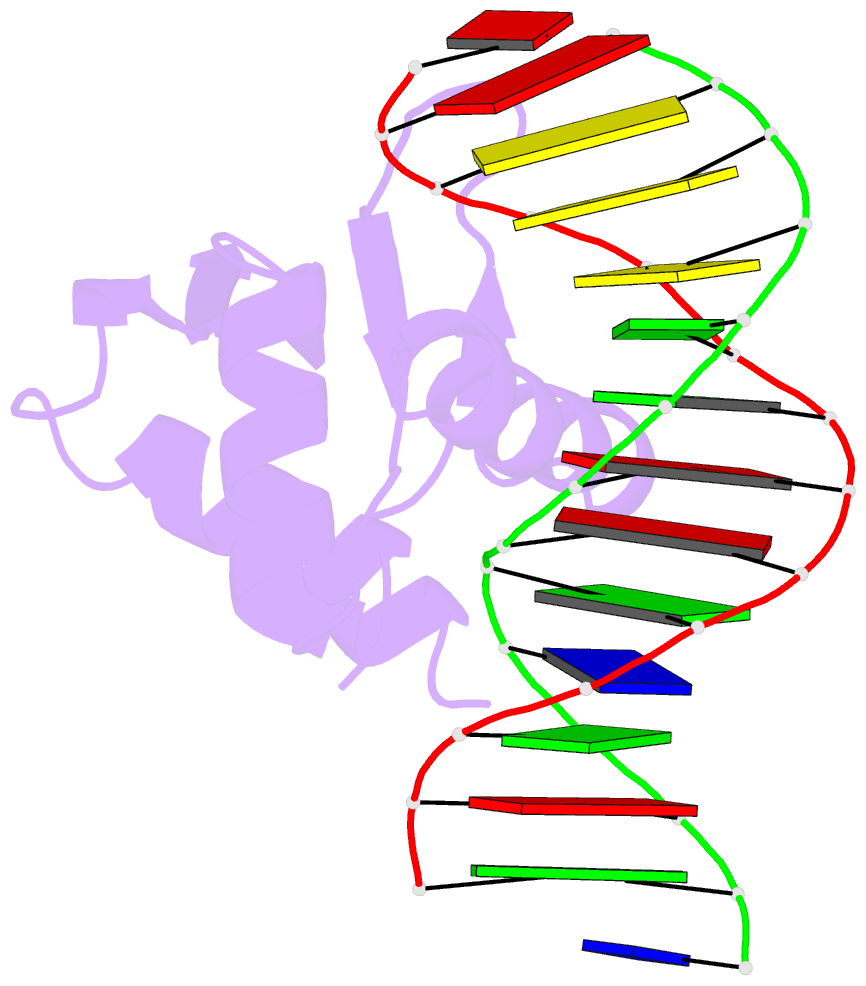
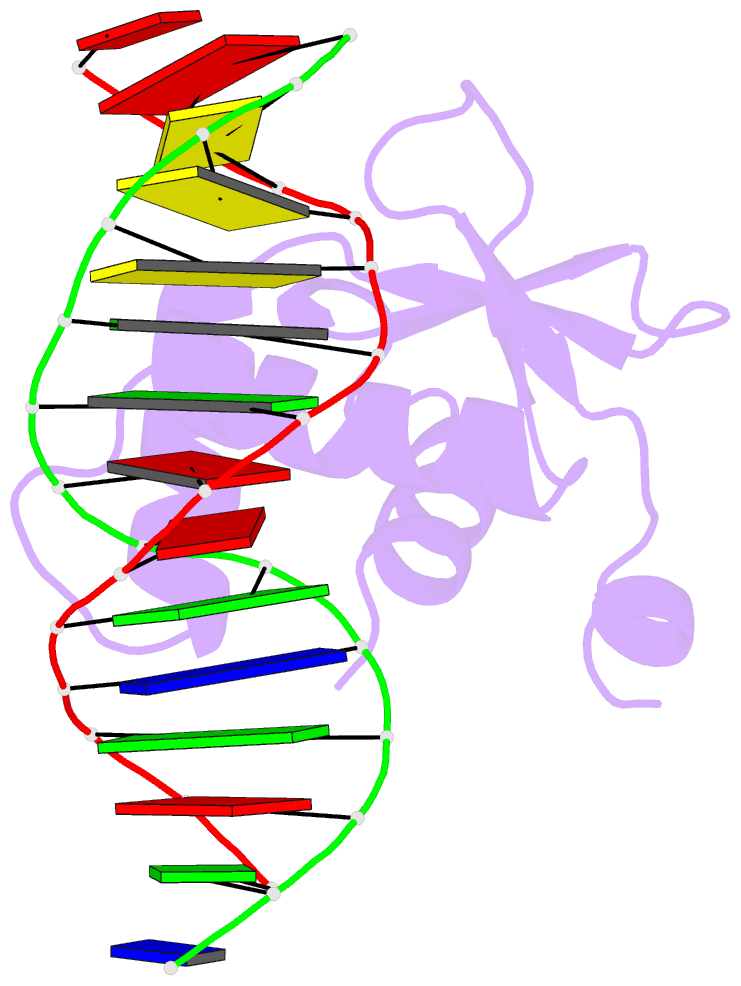
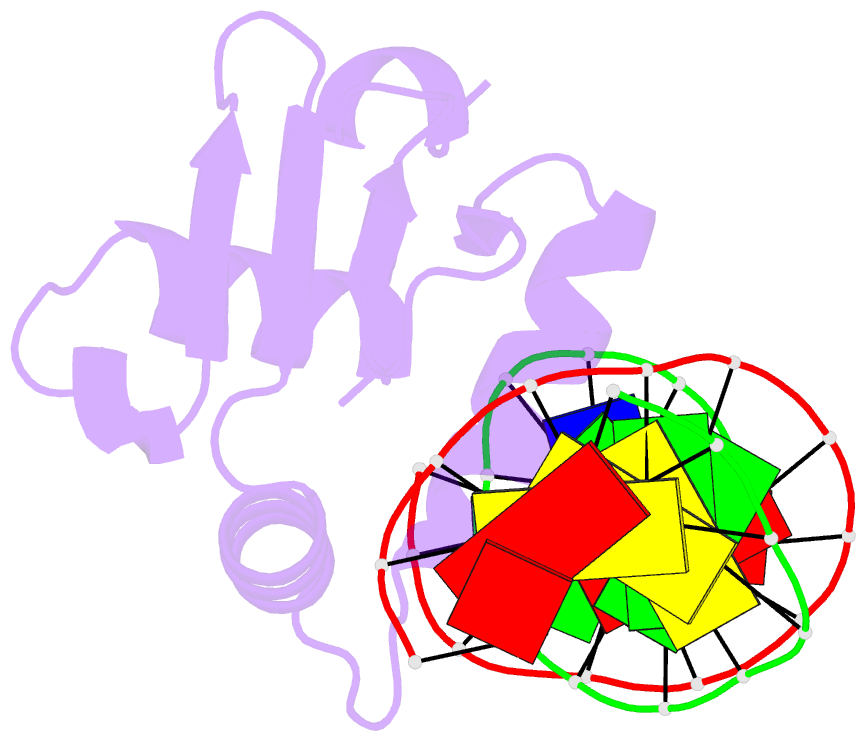
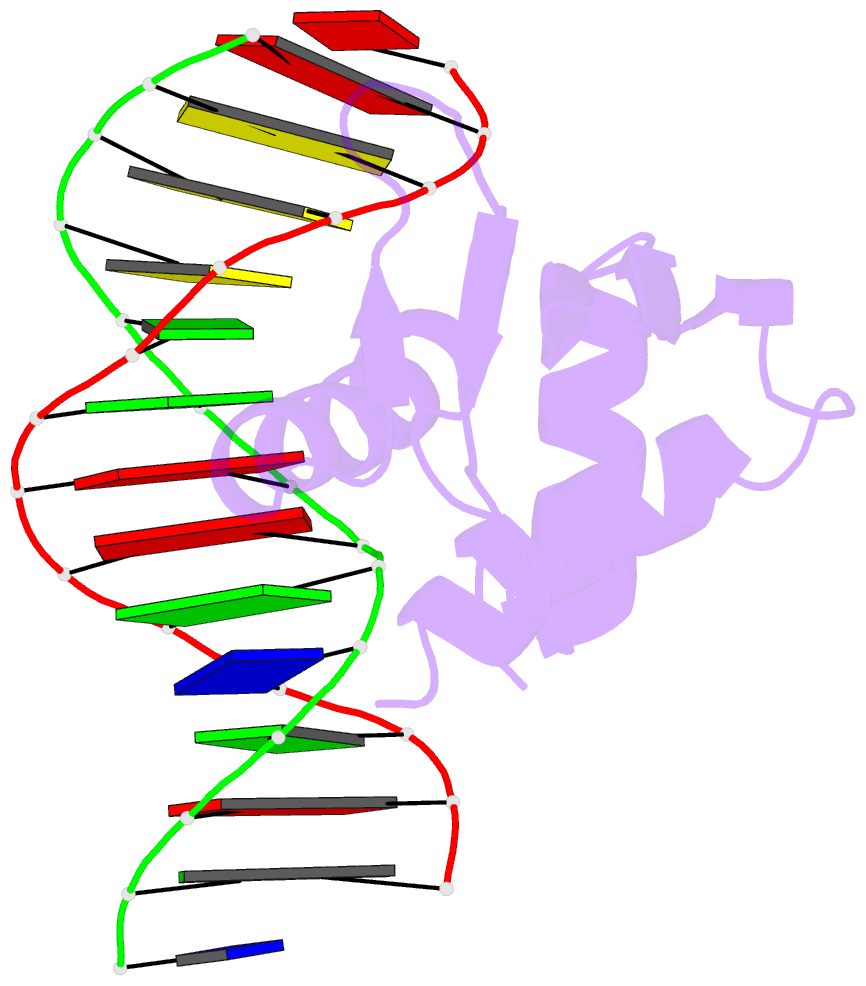
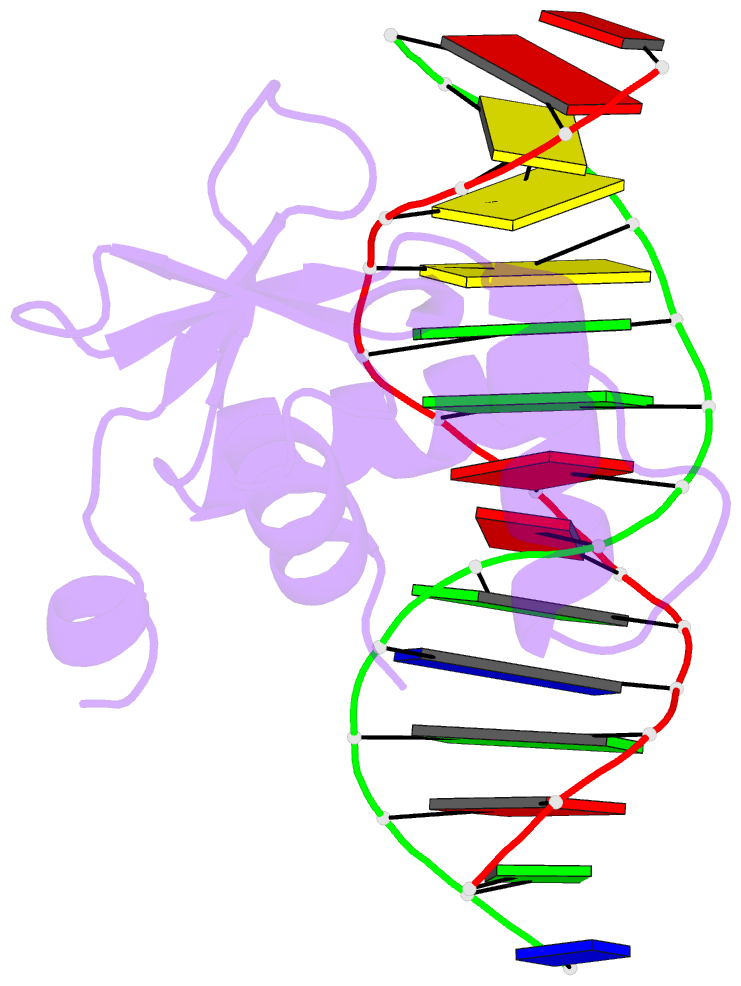
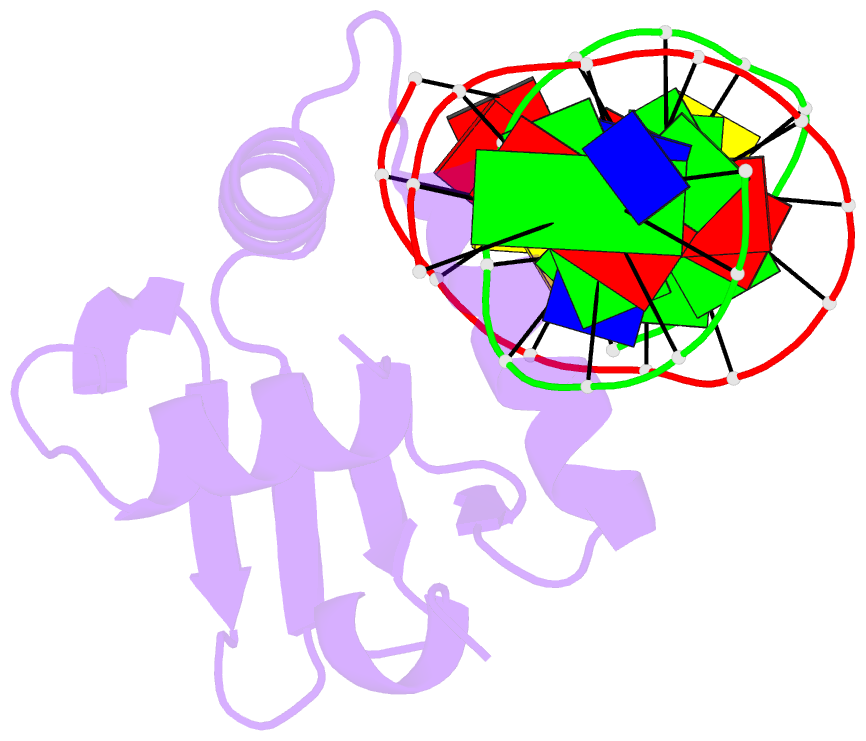
Summary information and primary citation
- PDB-id
- 4mhg; DSSR-derived features in text and JSON formats
- Class
- transcription-DNA
- Method
- X-ray (2.199 Å)
- Summary
- Crystal structure of etv6 bound to a specific DNA sequence
- Reference
- De S, Chan AC, Coyne HJ, Bhachech N, Hermsdorf U, Okon M, Murphy ME, Graves BJ, McIntosh LP (2014): "Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6." J.Mol.Biol., 426, 1390-1406. doi: 10.1016/j.jmb.2013.11.031.
- Abstract
- DNA binding by the ETS transcriptional repressor ETV6 (or TEL) is auto-inhibited~50-fold due to an α-helix that sterically blocks its ETS domain binding interface. Using NMR spectroscopy, we demonstrate that this marginally-stable helix is unfolded, and not displaced to a non-inhibitory position, when ETV6 is bound to DNA containing a consensus (5')GGAA(3') recognition site. Although significantly lower in affinity, binding to non-specific DNA is auto-inhibited~5-fold and also accompanied by helix unfolding. Based on NMR chemical shift perturbations, both specific and non-specific DNA are bound via the same canonical ETS domain interface. However, spectral perturbations are smaller for the non-specific complex, suggesting weaker and less well-defined interactions than in the specific complex. In parallel, the crystal structure of ETV6 bound to a specific DNA duplex was determined. The structure of this complex reveals that a non-conserved histidine residue in the ETS domain recognition helix helps establish the specificity of ETV6 for DNA-binding sites containing (5')GGAA(3') versus (5')GGAT(3'). These studies provide a unified steric mechanism for attenuating ETV6 binding to both specific and non-specific DNA and expand the repertoire of characterized auto-inhibitory strategies utilized to regulate ETS factors.