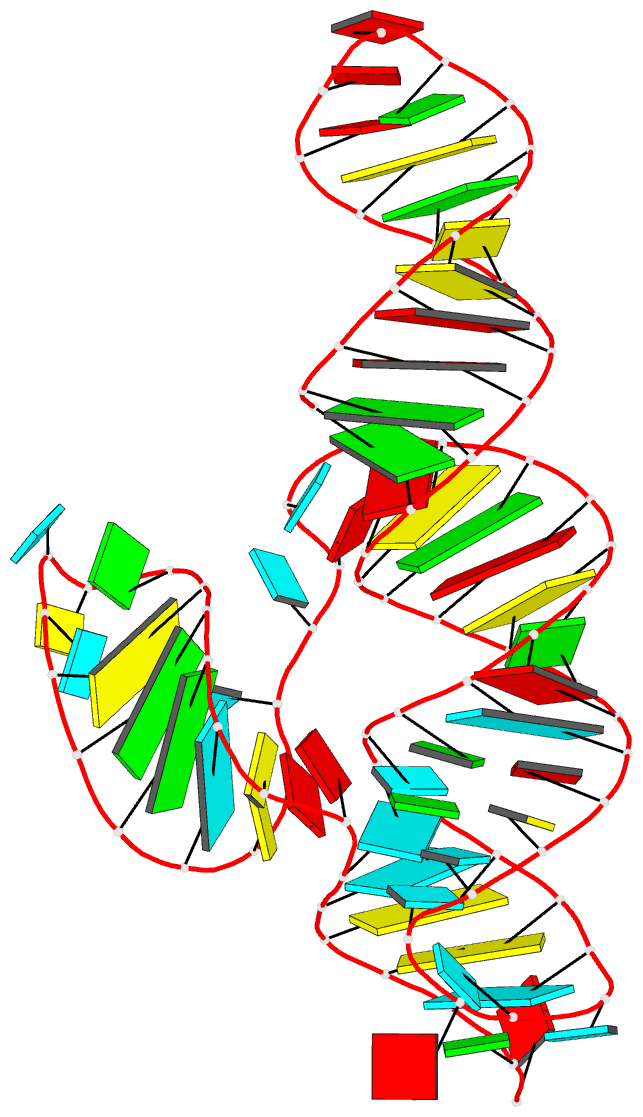
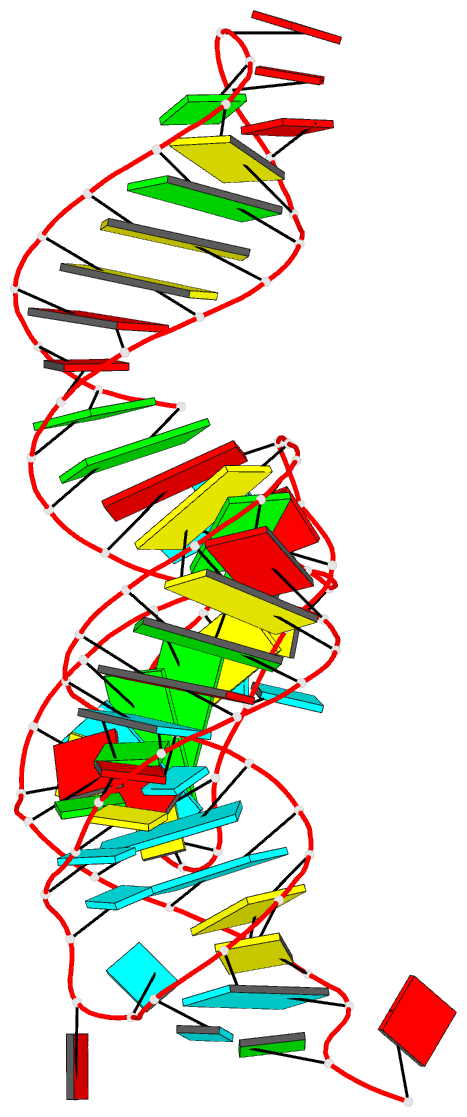
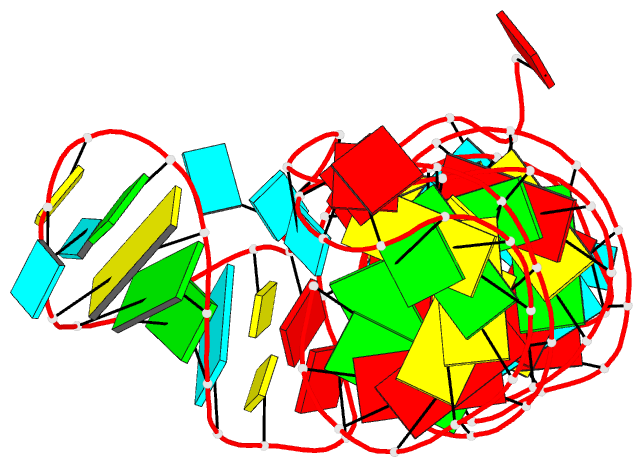
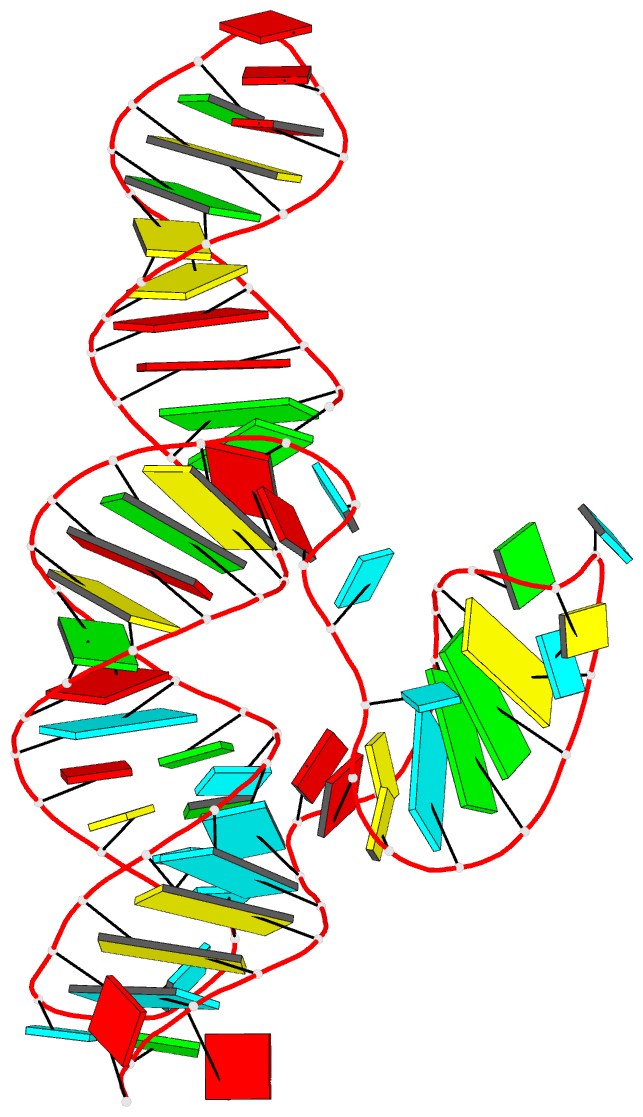
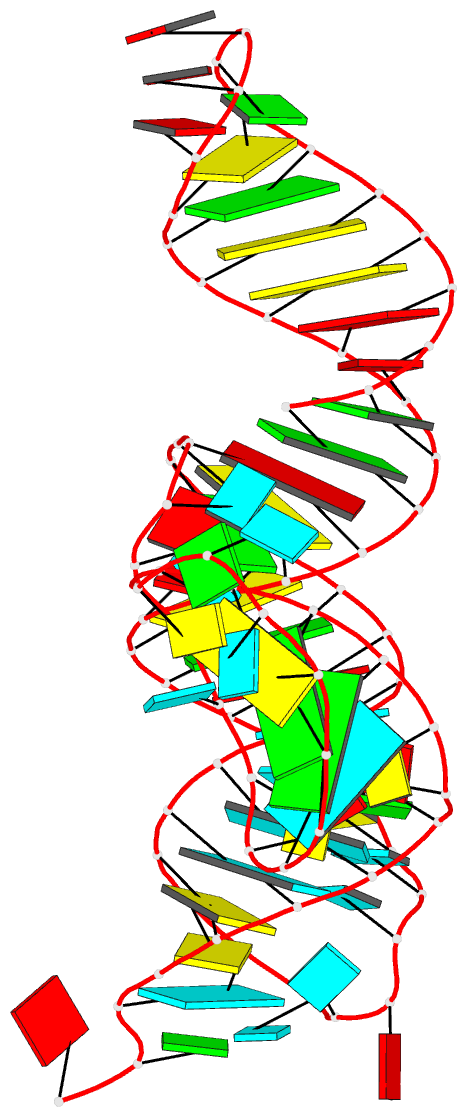
Summary information and primary citation
- PDB-id
- 4jf2; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.28 Å)
- Summary
- Structure of a class ii preq1 riboswitch reveals ligand recognition by a new fold
- Reference
- Liberman JA, Salim M, Krucinska J, Wedekind JE (2013): "Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold." Nat.Chem.Biol., 9, 353-355. doi: 10.1038/nchembio.1231.
- Abstract
- PreQ1 riboswitches regulate genes by binding the pyrrolopyrimidine intermediate preQ1 during the biosynthesis of the essential tRNA base queuosine. We report what is to our knowledge the first preQ1-II riboswitch structure at 2.3-Å resolution, which uses a previously uncharacterized fold to achieve effector recognition at the confluence of a three-way helical junction flanking a pseudoknotted ribosome-binding site. The results account for translational control mediated by the preQ1-II riboswitch class and expand the known repertoire of ligand-binding modes used by regulatory RNAs.