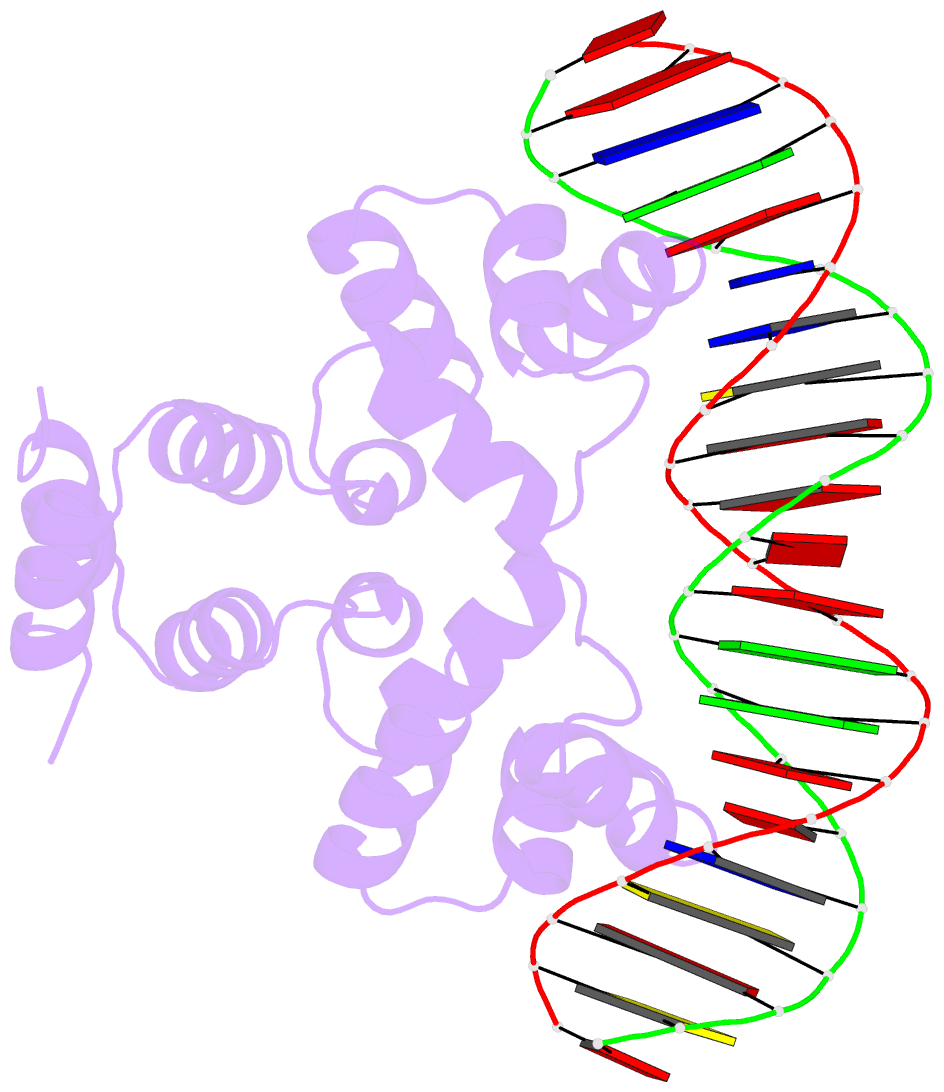
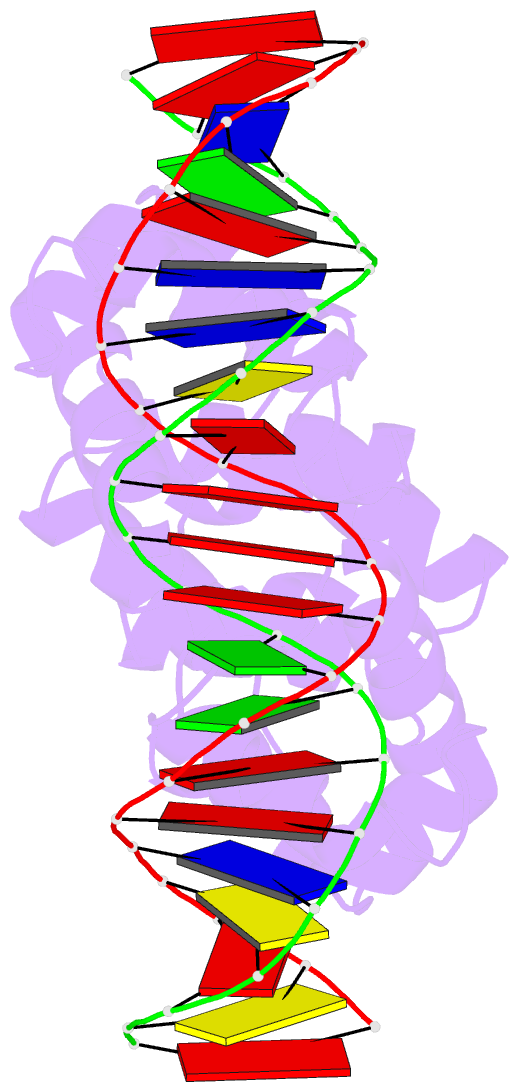
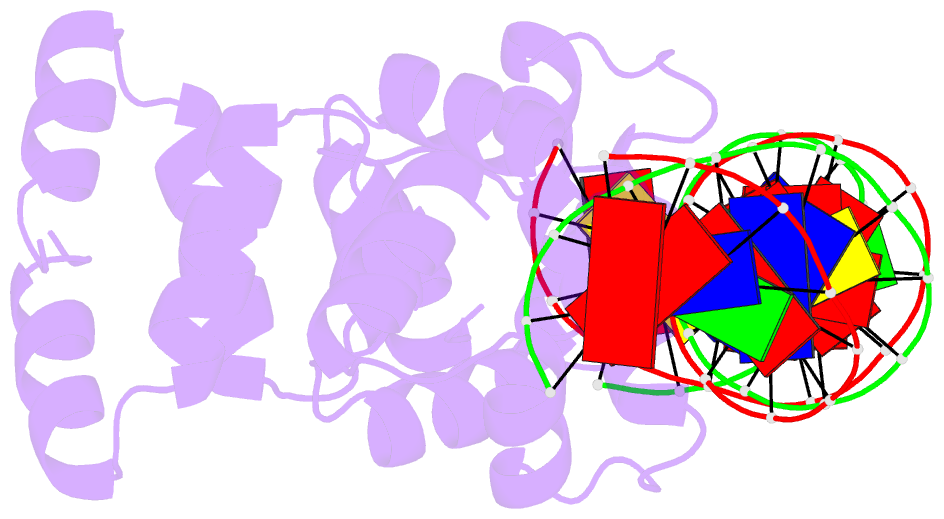
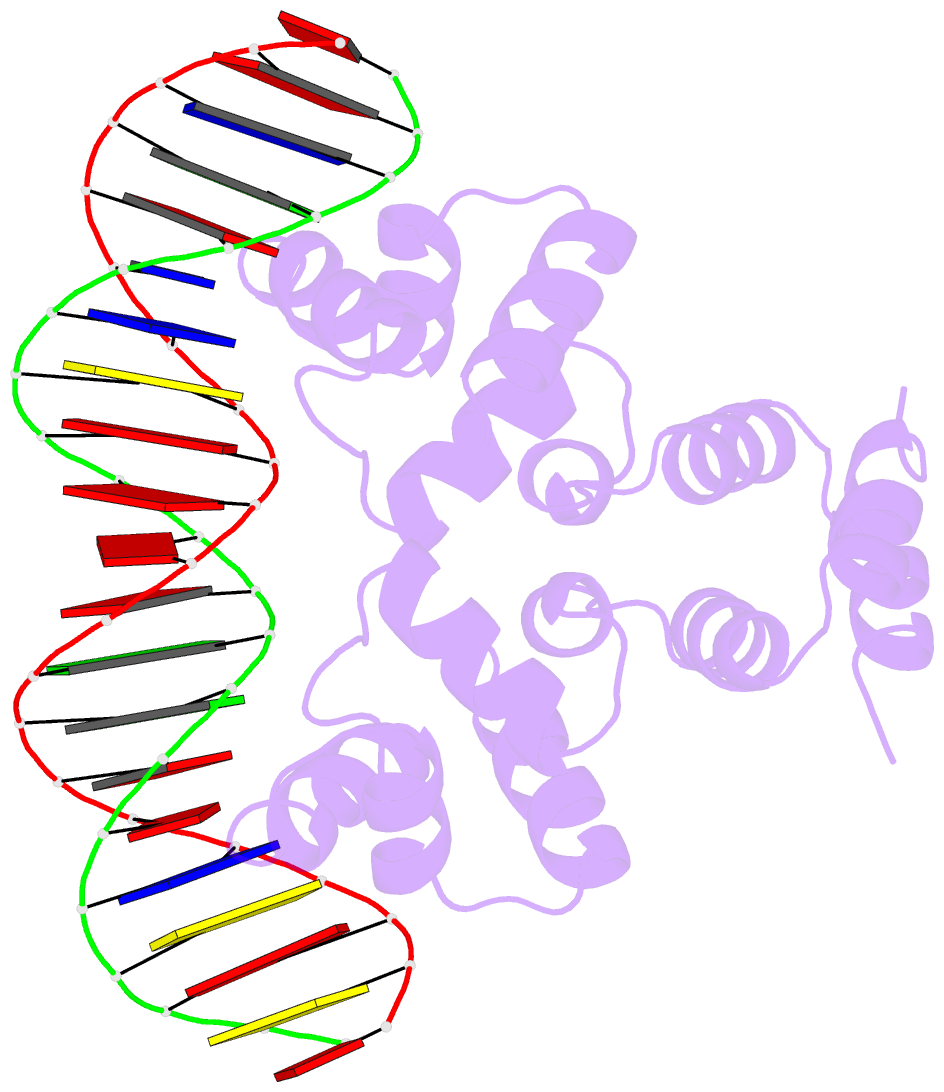
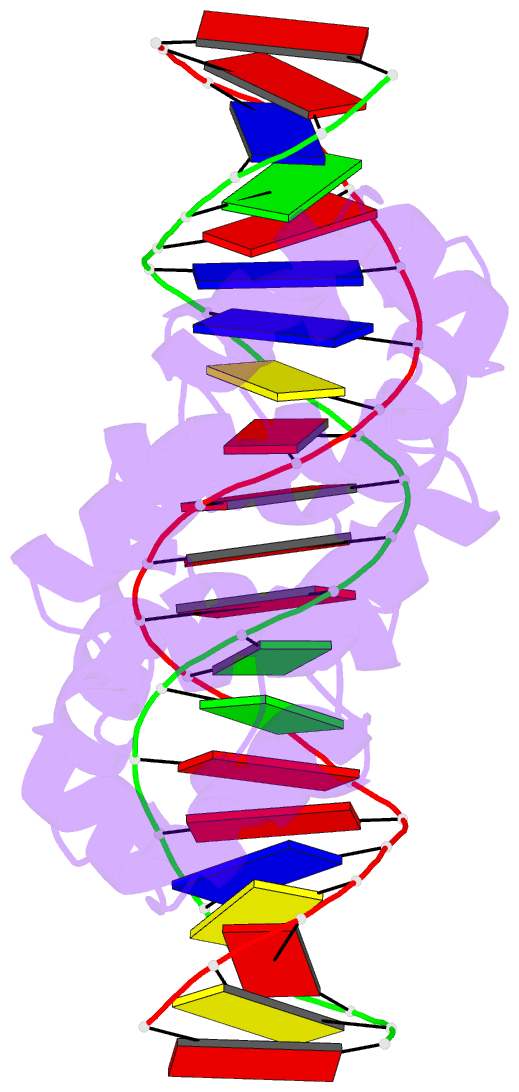
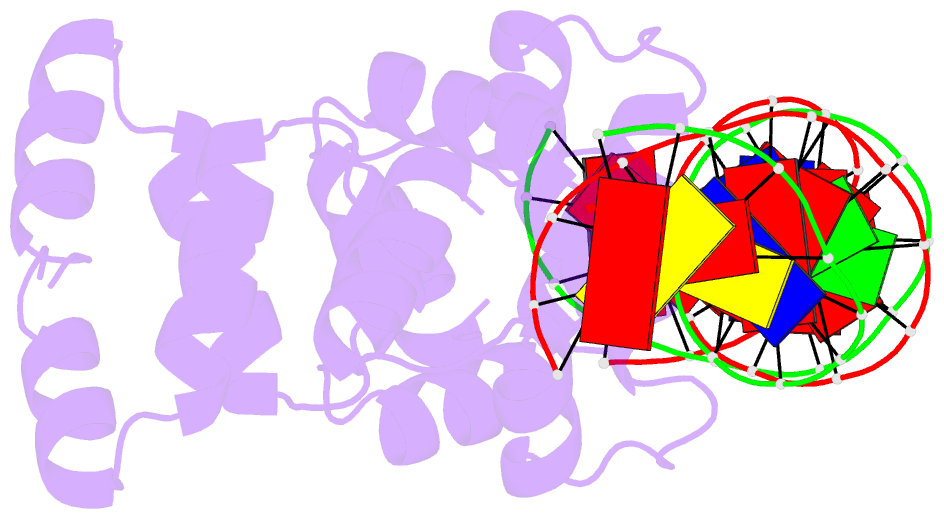
Summary information and primary citation
- PDB-id
- 4jcx; DSSR-derived features in text and JSON formats
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of the restriction-modification controller protein c.csp231i ol operator complex
- Reference
- Shevtsov MB, Streeter SD, Thresh SJ, Swiderska A, McGeehan JE, Kneale GG (2015): "Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression." Acta Crystallogr.,Sect.D, 71, 398-407. doi: 10.1107/S139900471402690X.
- Abstract
- In a wide variety of bacterial restriction-modification systems, a regulatory `controller' protein (or C-protein) is required for effective transcription of its own gene and for transcription of the endonuclease gene found on the same operon. We have recently turned our attention to a new class of controller proteins (exemplified by C.Csp231I) that have quite novel features, including a much larger DNA-binding site with an 18 bp (∼60 Å) spacer between the two palindromic DNA-binding sequences and a very different recognition sequence from the canonical GACT/AGTC. Using X-ray crystallography, the structure of the protein in complex with its 21 bp DNA-recognition sequence was solved to 1.8 Å resolution, and the molecular basis of sequence recognition in this class of proteins was elucidated. An unusual aspect of the promoter sequence is the extended spacer between the dimer binding sites, suggesting a novel interaction between the two C-protein dimers when bound to both recognition sites correctly spaced on the DNA. A U-bend model is proposed for this tetrameric complex, based on the results of gel-mobility assays, hydrodynamic analysis and the observation of key contacts at the interface between dimers in the crystal.