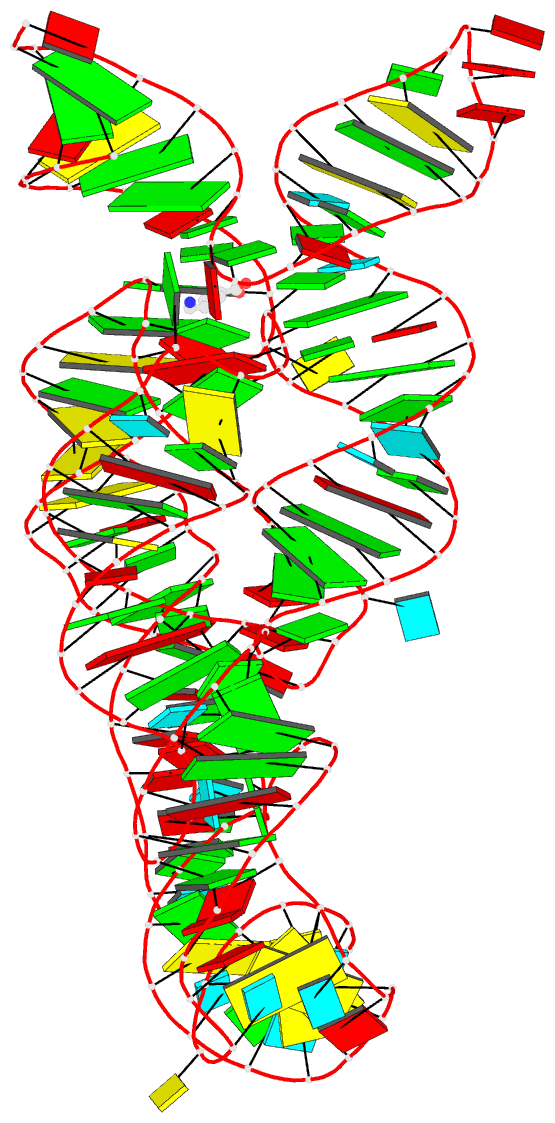
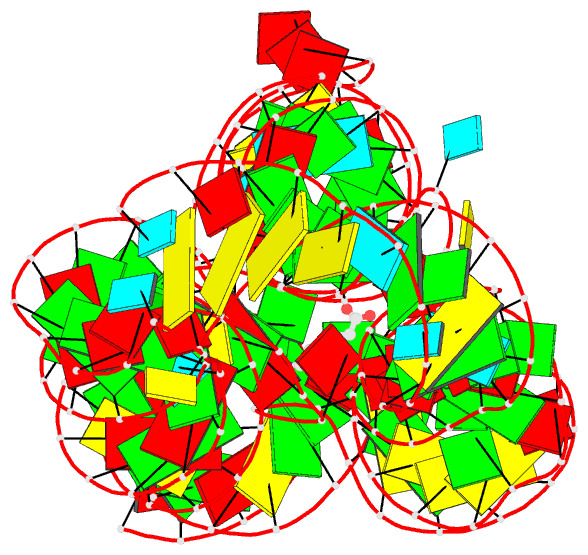
Summary information and primary citation
- PDB-id
- 4erj; DSSR-derived features in text and JSON formats
- Class
- transcription
- Method
- X-ray (3.0 Å)
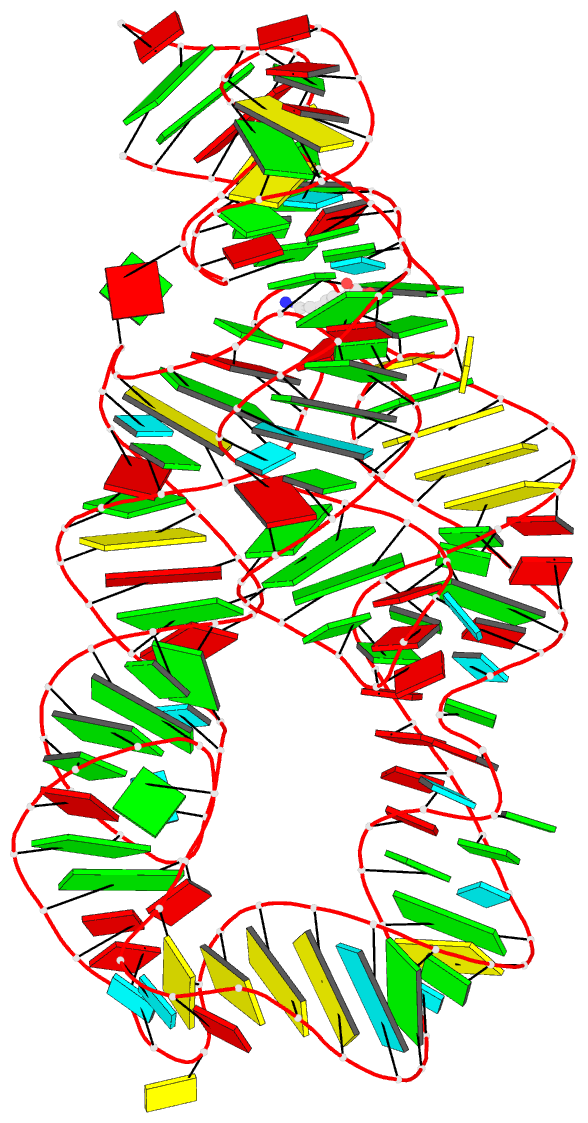
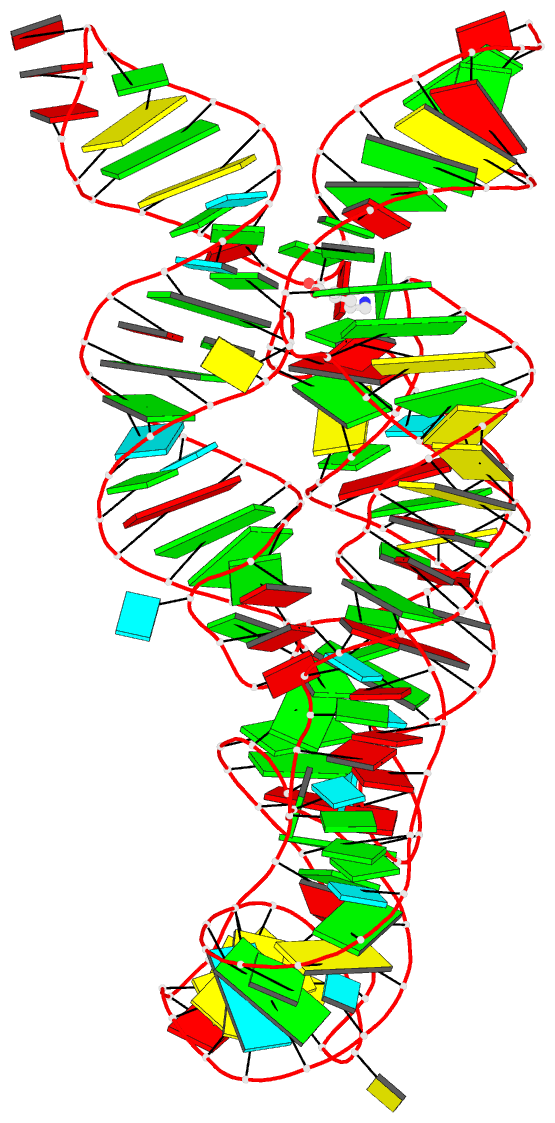
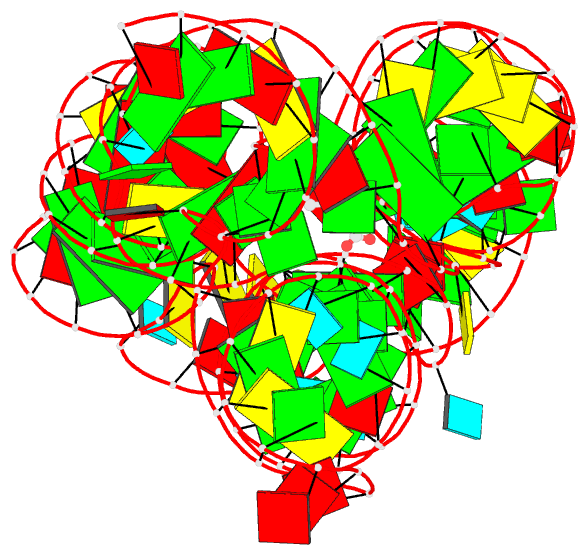
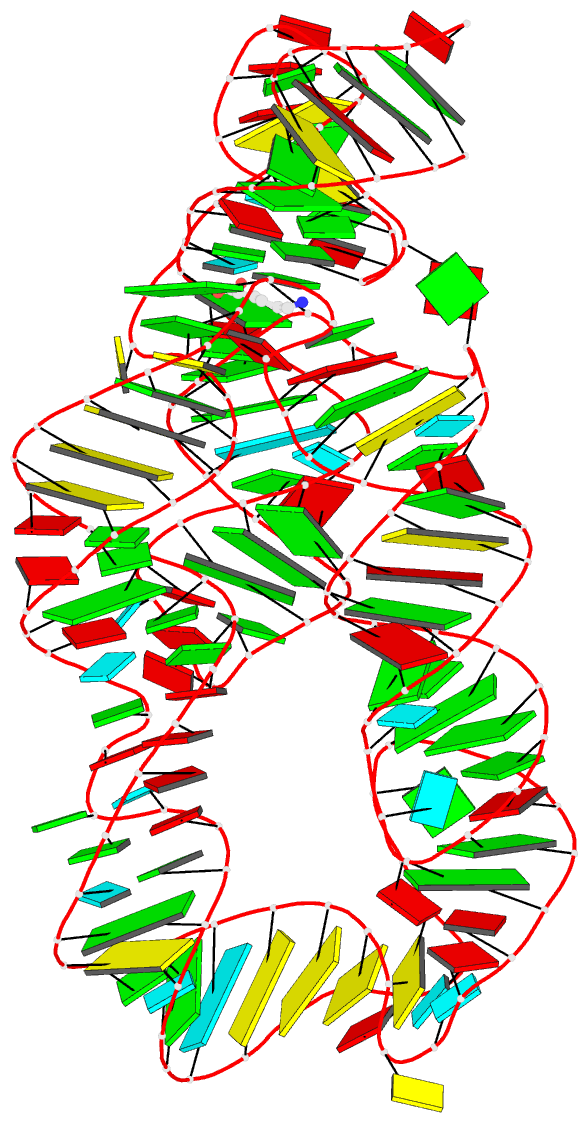
- Summary
- Crystal structure of the lysine riboswitch bound to a 6-aminocaproic acid
- Reference
- Garst AD, Porter EB, Batey RT (2012): "Insights into the regulatory landscape of the lysine riboswitch." J.Mol.Biol., 423, 17-33. doi: 10.1016/j.jmb.2012.06.038.
- Abstract
- A prevalent means of regulating gene expression in bacteria is by riboswitches found within mRNA leader sequences. Like protein repressors these RNA elements must bind an effector molecule with high specificity against a background of other cellular metabolites of similar chemical structure to elicit the appropriate regulatory response. Current crystal structures of the lysine riboswitch do not provide a complete understanding of selectivity as recognition is substantially mediated through main chain atoms of the amino acid. Using a directed set of lysine analogs and other amino acids, the relative contributions of the polar functional groups to binding affinity and the regulatory response have been determined. Our results reveal that the lysine riboswitch has >1,000-fold specificity for lysine over other amino acids. To achieve this specificity, the aptamer is highly sensitive to the precise placement of the ε-amino group and relatively tolerant of alterations to the main chain functional groups. At low NTP concentrations, we observe good agreement between the half-maximal regulatory activity (T(50)) and the affinity of the receptor for lysine (K(D)) as well many of its analogs. However, above 400 μM [NTP] the concentration of lysine required to elicit transcription termination rises, moving into the riboswitch into a kinetic control regime. These data demonstrate that under physiologically relevant conditions riboswitches can integrate both effector and NTP concentrations to generate a regulatory response appropriate for global metabolic state of the cell.