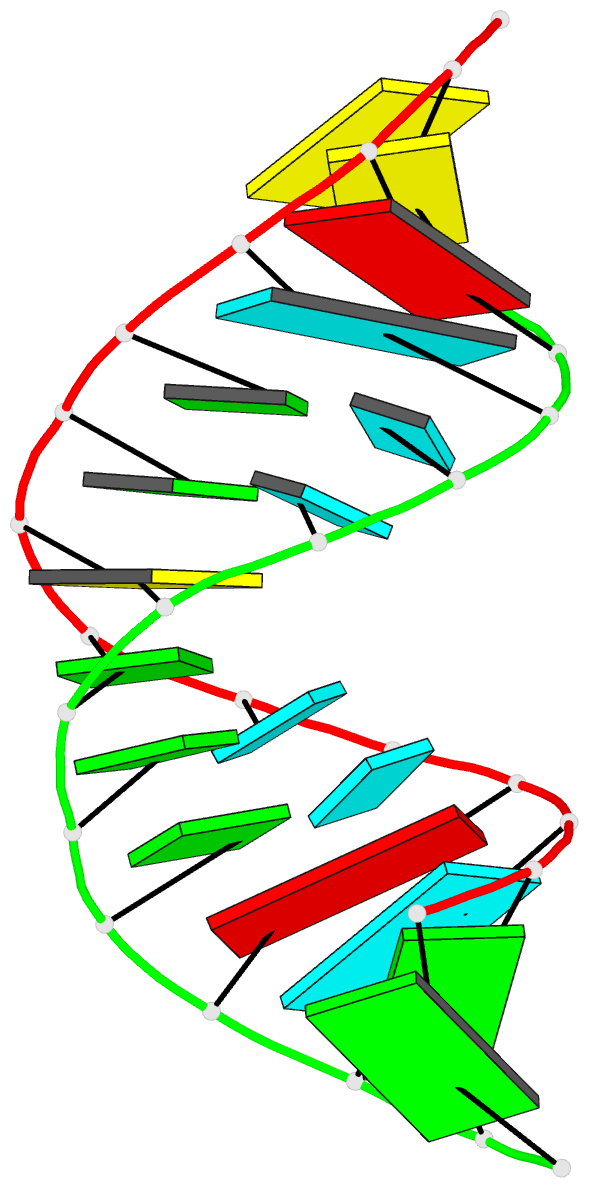
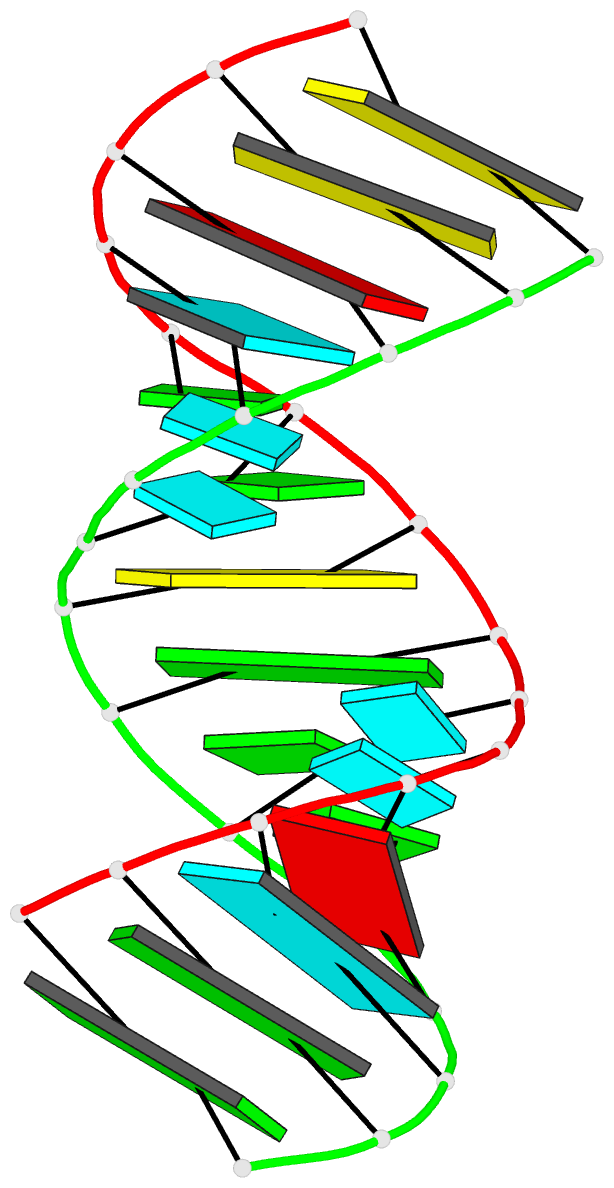
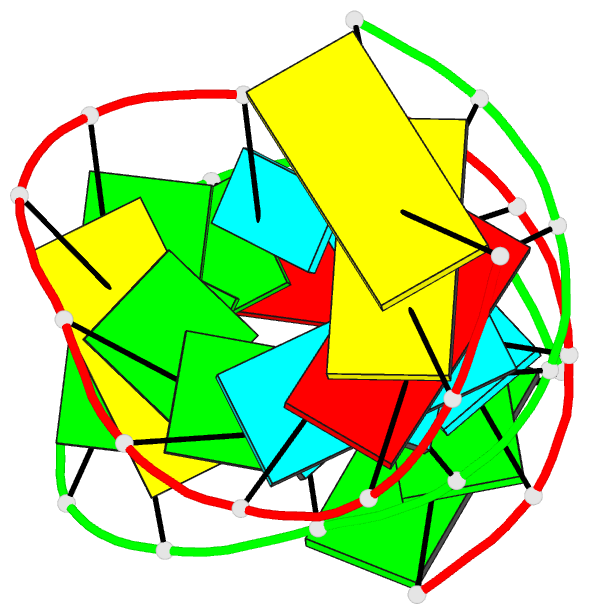
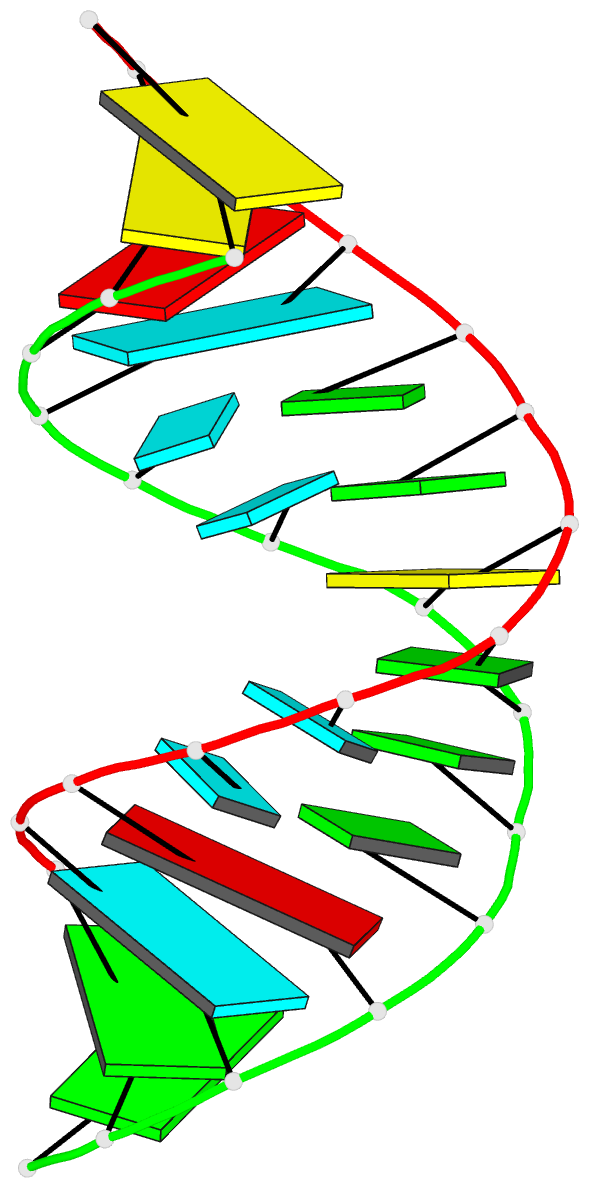
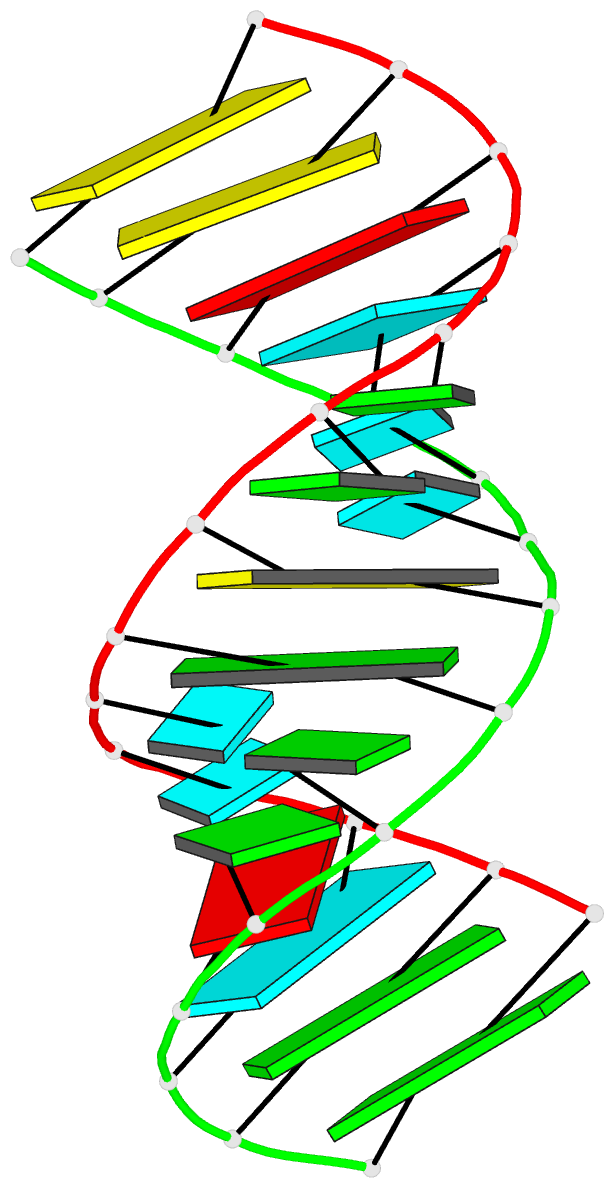
Summary information and primary citation
- PDB-id
- 433d; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.1 Å)
- Summary
- Crystal structure of a 14 base pair RNA duplex with nonsymmetrical tandem g.u wobble base pairs
- Reference
- Trikha J, Filman DJ, Hogle JM (1999): "Crystal structure of a 14 bp RNA duplex with non-symmetrical tandem GxU wobble base pairs." Nucleic Acids Res., 27, 1728-1739. doi: 10.1093/nar/27.7.1728.
- Abstract
- Adjacent GxU wobble base pairs are frequently found in rRNA. Atomic structures of small RNA motifs help to provide a better understanding of the effects of various tandem mismatches on duplex structure and stability, thereby providing better rules for RNA structure prediction and validation. The crystal structure of an RNA duplex containing the sequence r(GGUAUUGC-GGUACC)2 has been solved at 2.1 A resolution using experimental phases. Novel refinement strategies were needed for building the correct solvent model. At present, this is the only short RNA duplex structure containing 5'-U-U-3'/3'-G-G-5' non-symmetric tandem GxU wobble base pairs. In the 14mer duplex, the six central base pairs are all displaced away from the helix axis, yielding significant changes in local backbone conformation, helix parameters and charge distribution that may provide specific recognition sites for biologically relevant ligand binding. The greatest deviations from A-form helix occur where the guanine of a wobble base pair stacks over a purine from the opposite strand. In this vicinity, the intra-strand phosphate distances increase significantly, and the major groove width increases up to 3 A. Structural comparisons with other short duplexes containing symmetrical tandem GxU or GxT wobble base pairs show that nearest-neighbor sequence dependencies govern helical twist and the occurrence of cross-strand purine stacks.