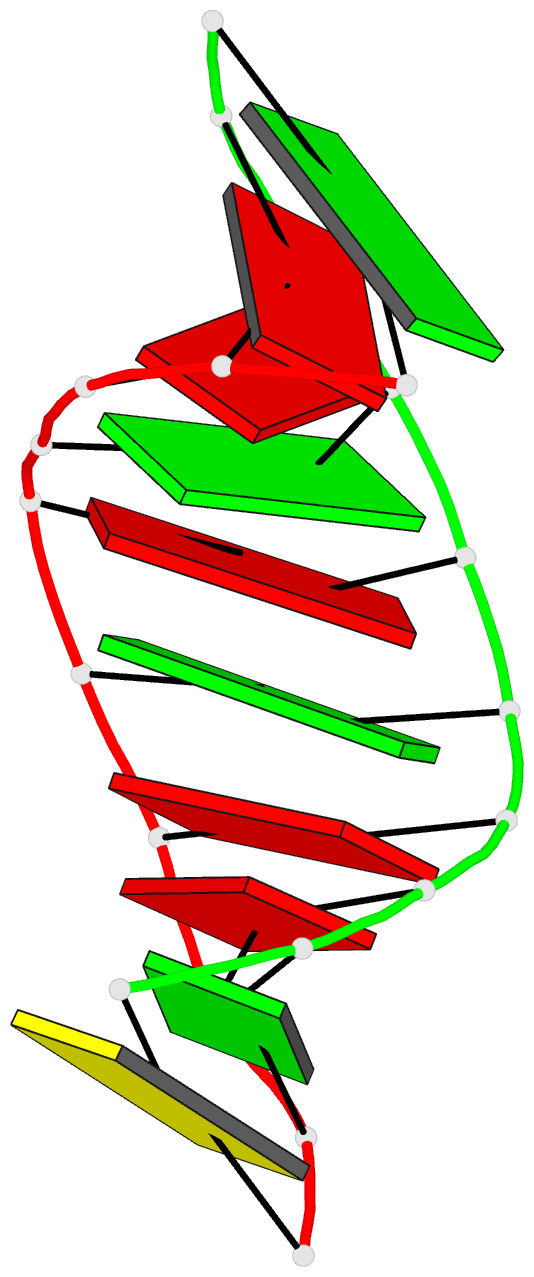
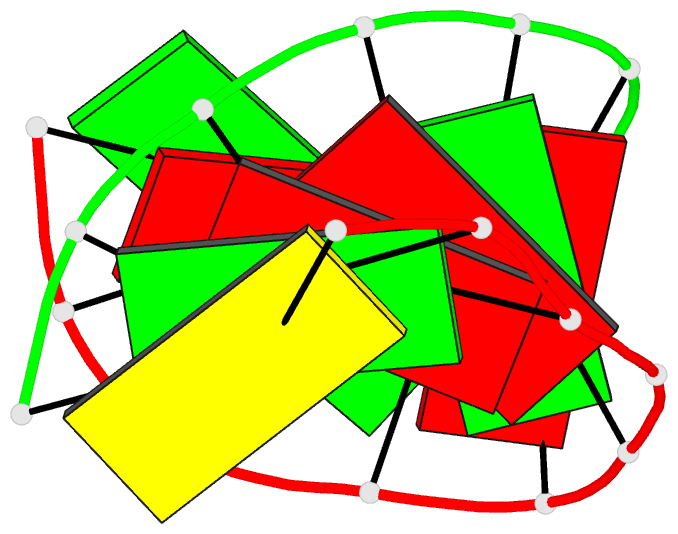
Summary information and primary citation
- PDB-id
- 404d; DSSR-derived features in text and JSON formats
- Class
- DNA-RNA hybrid
- Method
- X-ray (2.5 Å)
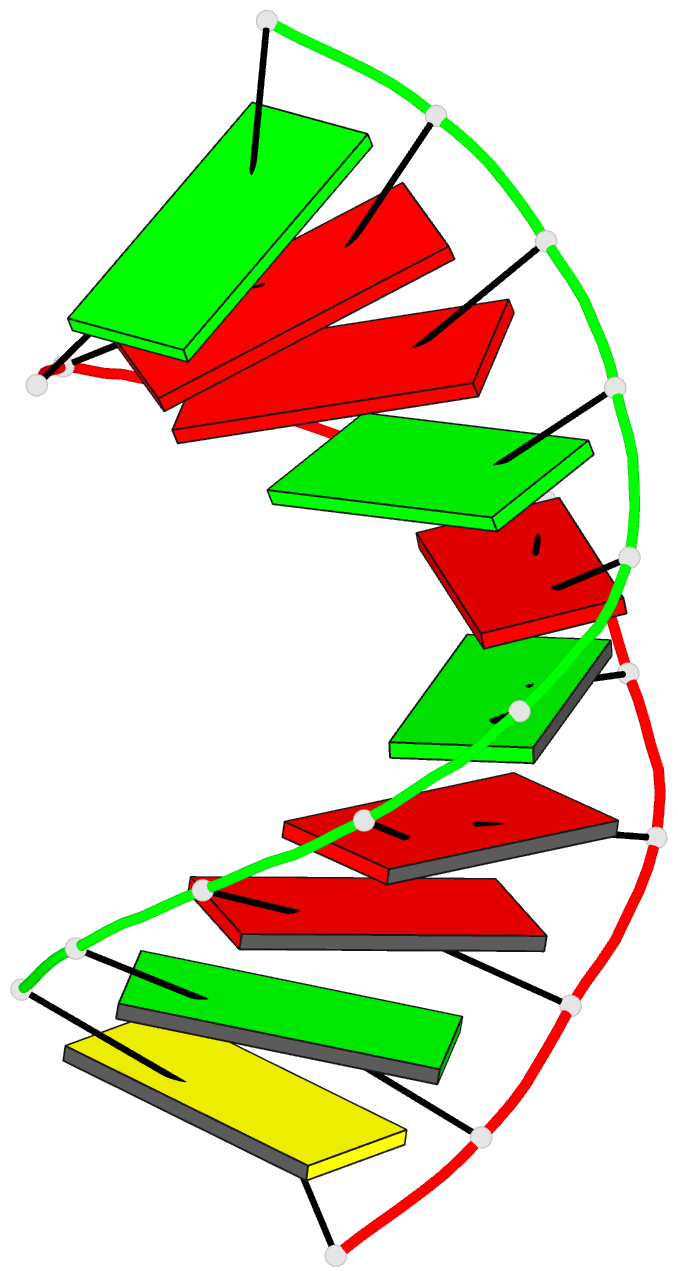
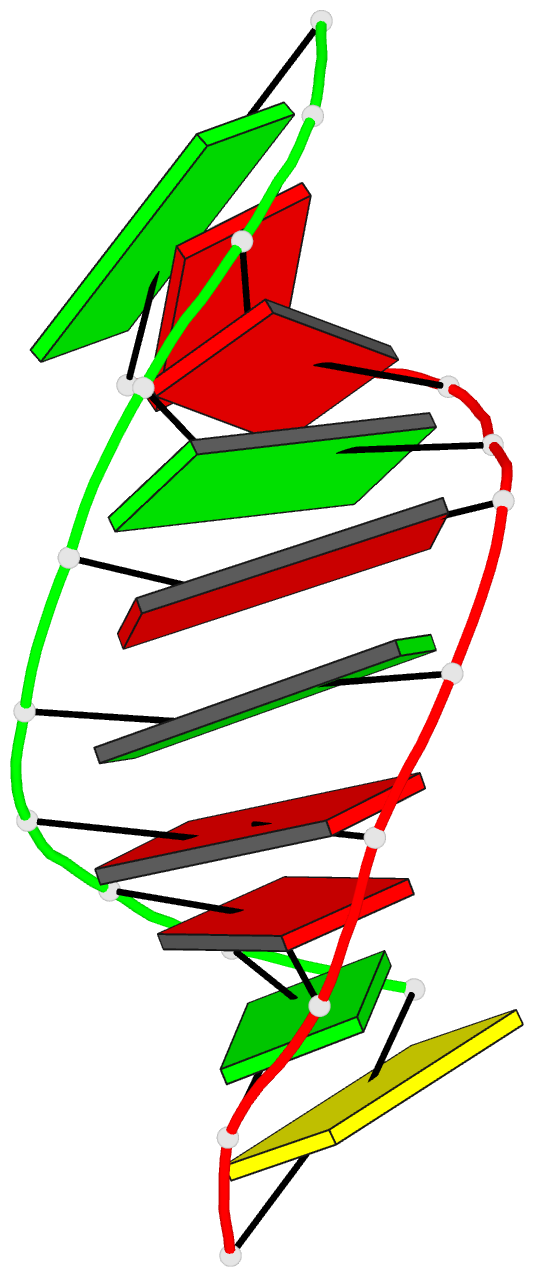
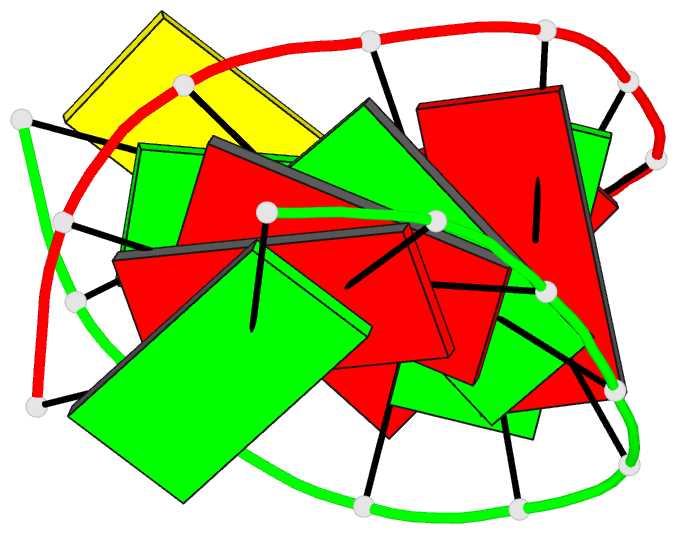
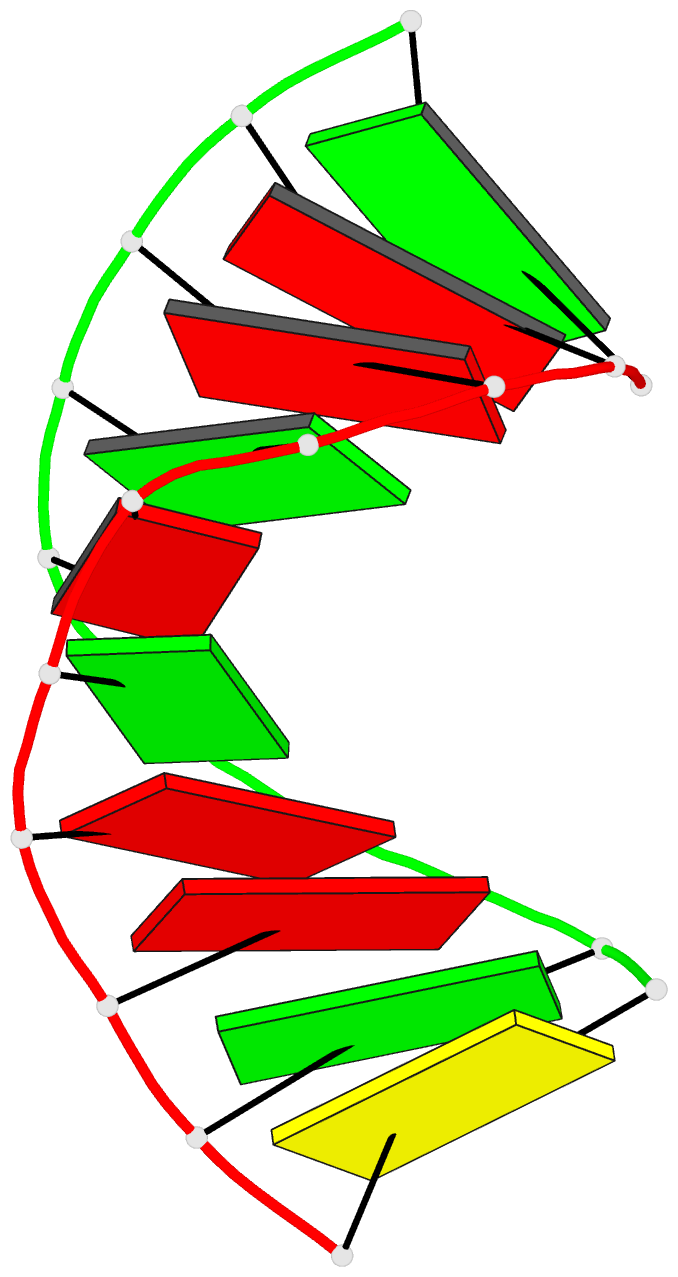
- Summary
- Crystal structure of the RNA-DNA hybrid r(gaagagaagc). d(gcttctcttc)
- Reference
- Conn GL, Brown T, Leonard GA (1999): "The crystal structure of the RNA/DNA hybrid r(GAAGAGAAGC). d(GCTTCTCTTC) shows significant differences to that found in solution." Nucleic Acids Res., 27, 555-561. doi: 10.1093/nar/27.2.555.
- Abstract
- The crystal structure of the RNA/DNA hybrid r(GAAGAGAAGC). d(GCTTCTCTTC) has been solved and refined at 2.5 A resolution. The refinement procedure converged at R = 0.181 for all reflections in the range 20.0-2.5 A. In the crystal, the RNA/DNA hybrid duplex has an A' conformation with all but one of the nucleotide sugar moieties adopting a C3'- endo (N) conformation. Both strands in the double helix adopt a global conformation close to the A-form and the width of the minor groove is typical of that found in the crystal structures of other A-form duplexes. However, differences are observed between the RNA and DNA strands that make up the hybrid at the local level. In the central portion of the duplex, the RNA strand has backbone alpha, beta and gamma torsion angles that alternate between the normal gauche -/ trans / gauche + conformation and an unusual trans / trans / trans conformation. Coupled with this so-called 'alpha/gamma flipping' of the backbone torsion angles, the distance between adjacent phosphorous atoms on the RNA strand systematically varies. Neither of these phenomena are observed on the DNA strand. The structure of the RNA/DNA hybrid presented here differs significantly from that found in solution for this and other sequences. Possible reasons for these differences and their implications for the current model of RNase H activity are discussed.