Summary information and primary citation
- PDB-id
-
3sux;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (2.9 Å)
- Summary
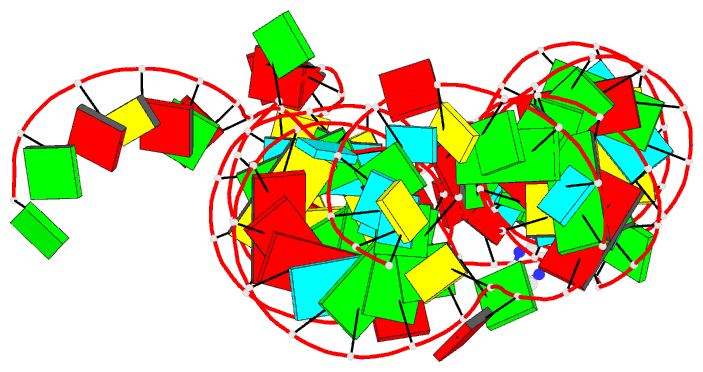
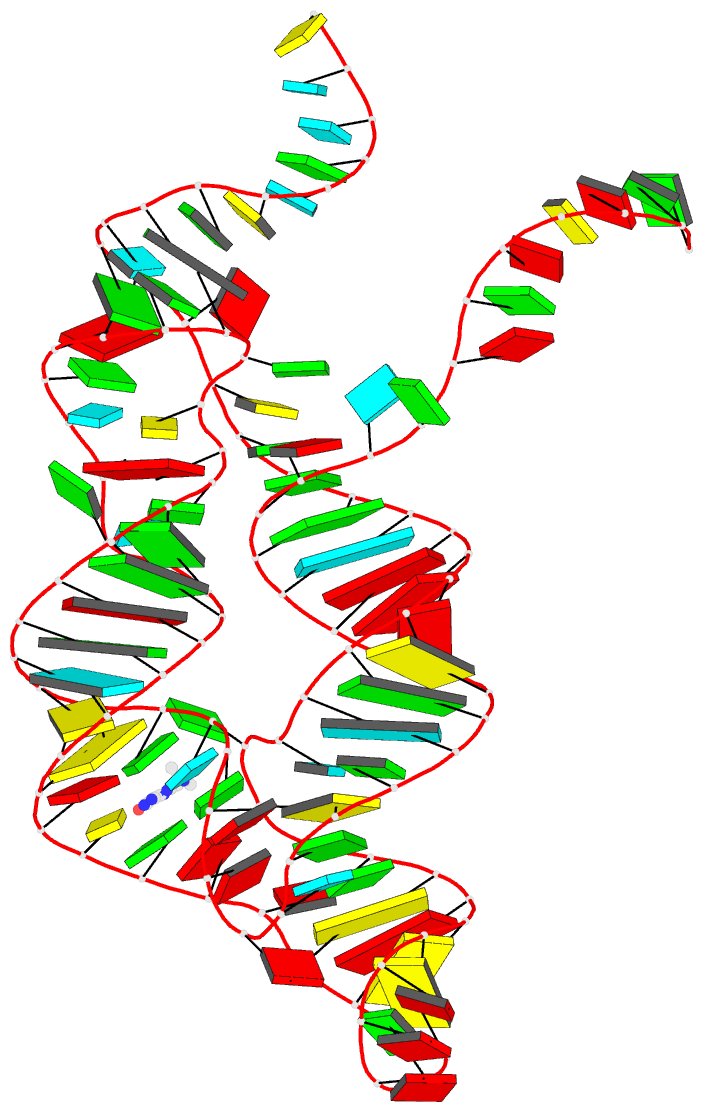
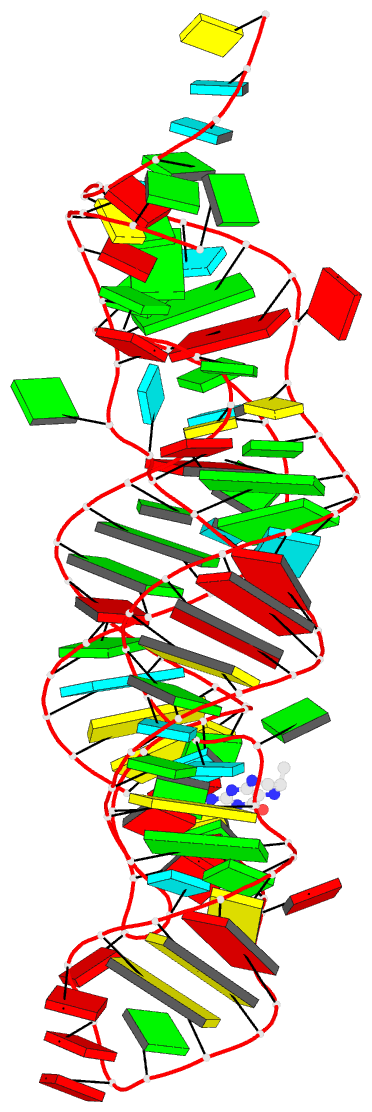
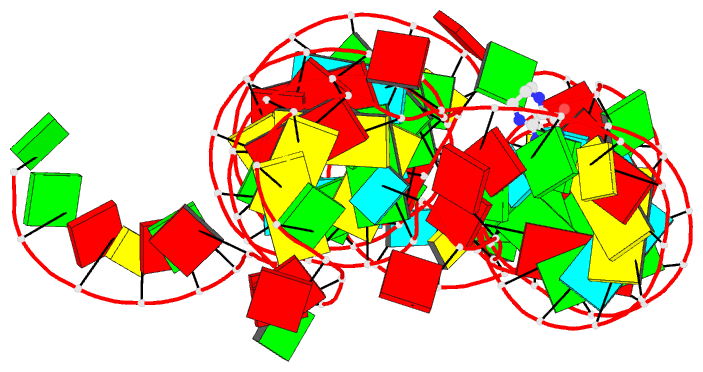
- Crystal structure of thf riboswitch, bound with
thf
- Reference
-
Huang L, Ishibe-Murakami S, Patel DJ, Serganov A (2011):
"Long-range
pseudoknot interactions dictate the regulatory response
in the tetrahydrofolate riboswitch."
Proc.Natl.Acad.Sci.USA, 108,
14801-14806. doi: 10.1073/pnas.1111701108.
- Abstract
- Tetrahydrofolate (THF), a biologically active form of
the vitamin folate (B(9)), is an essential cofactor in
one-carbon transfer reactions. In bacteria, expression of
folate-related genes is controlled by feedback modulation
in response to specific binding of THF and related
compounds to a riboswitch. Here, we present the X-ray
structures of the THF-sensing domain from the Eubacterium
siraeum riboswitch in the ligand-bound and unbound states.
The structure reveals an "inverted" three-way junctional
architecture, most unusual for riboswitches, with the
junction located far from the regulatory helix P1 and not
directly participating in helix P1 formation. Instead, the
three-way junction, stabilized by binding to the ligand,
aligns the riboswitch stems for long-range tertiary
pseudoknot interactions that contribute to the organization
of helix P1 and therefore stipulate the regulatory response
of the riboswitch. The pterin moiety of the ligand docks in
a semiopen pocket adjacent to the junction, where it forms
specific hydrogen bonds with two moderately conserved
pyrimidines. The aminobenzoate moiety stacks on a guanine
base, whereas the glutamate moiety does not appear to make
strong interactions with the RNA. In contrast to other
riboswitches, these findings demonstrate that the THF
riboswitch uses a limited number of available determinants
for ligand recognition. Given that modern antibiotics
target folate metabolism, the THF riboswitch structure
provides insights on mechanistic aspects of riboswitch
function and may help in manipulating THF levels in
pathogenic bacteria.