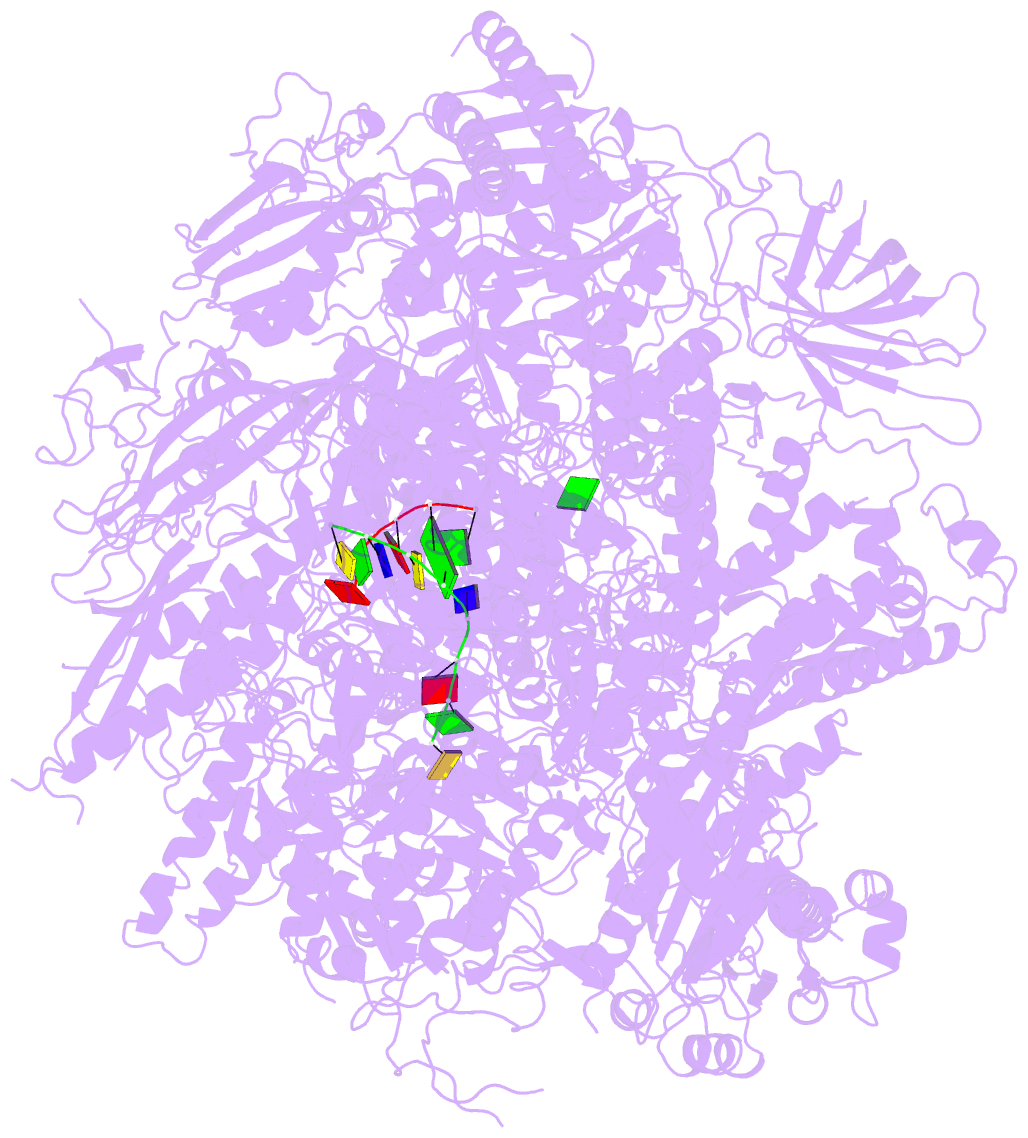
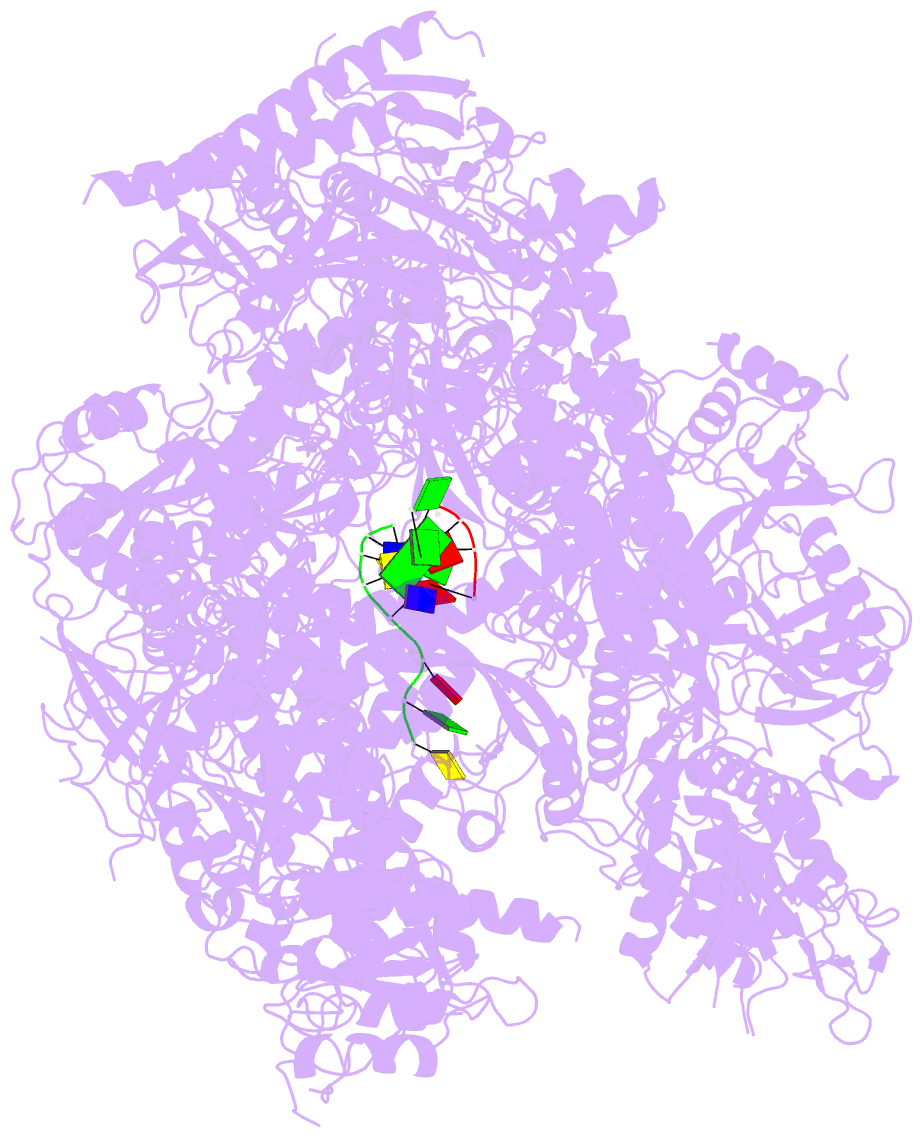
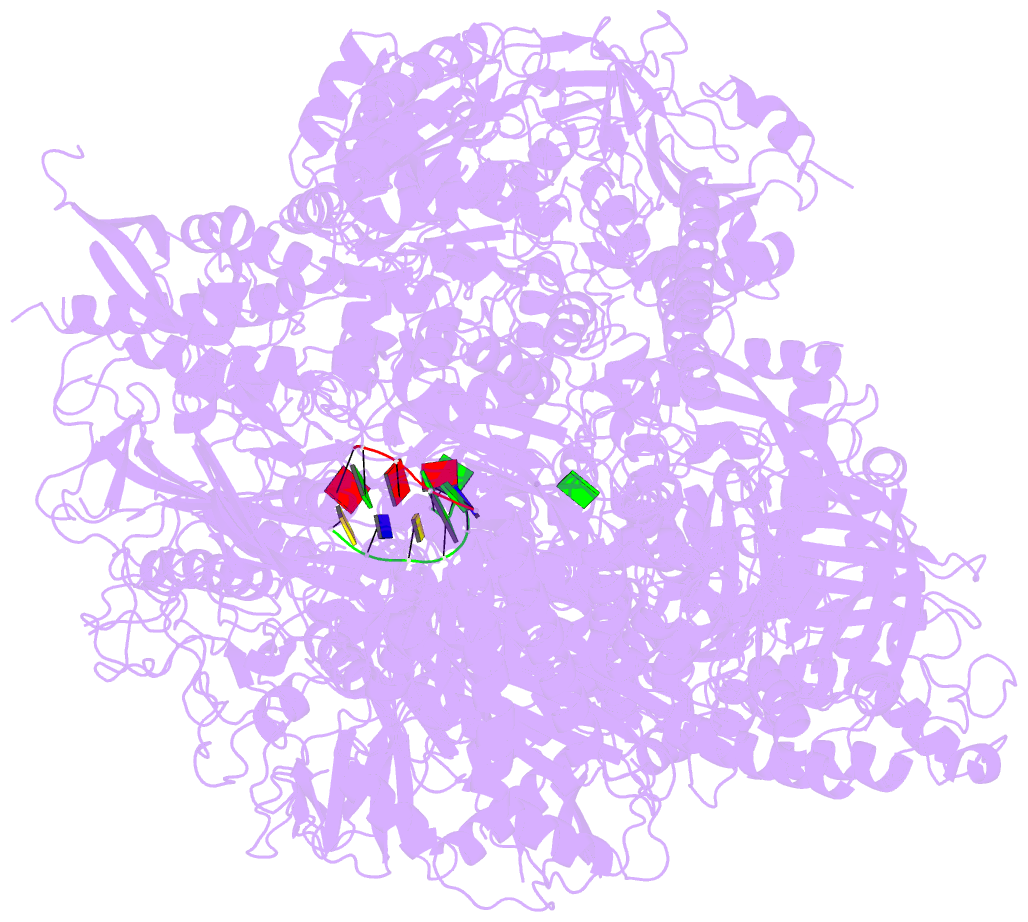
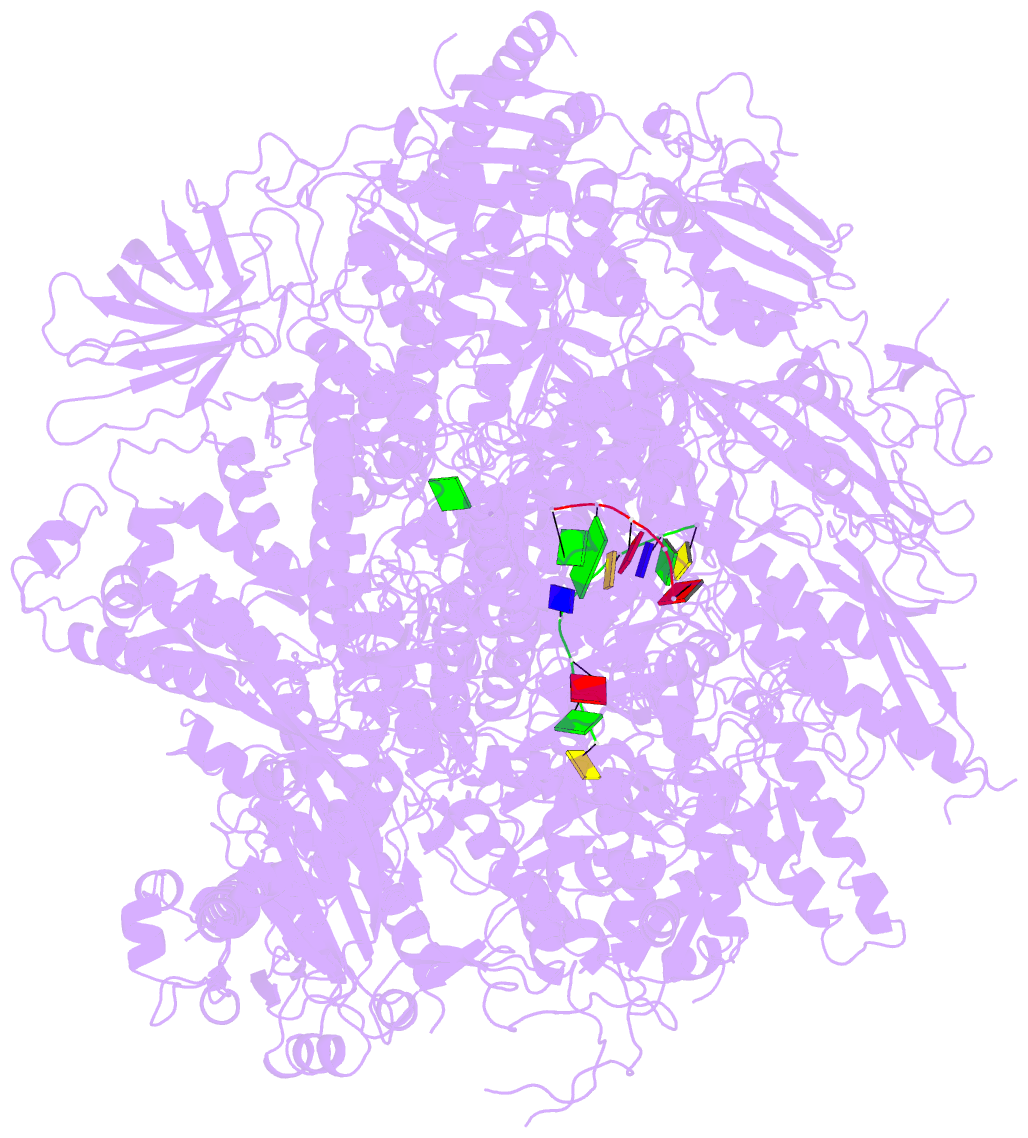
Summary information and primary citation
- PDB-id
- 3s1r; DSSR-derived features in text and JSON formats
- Class
- transcription-RNA-DNA
- Method
- X-ray (3.2 Å)
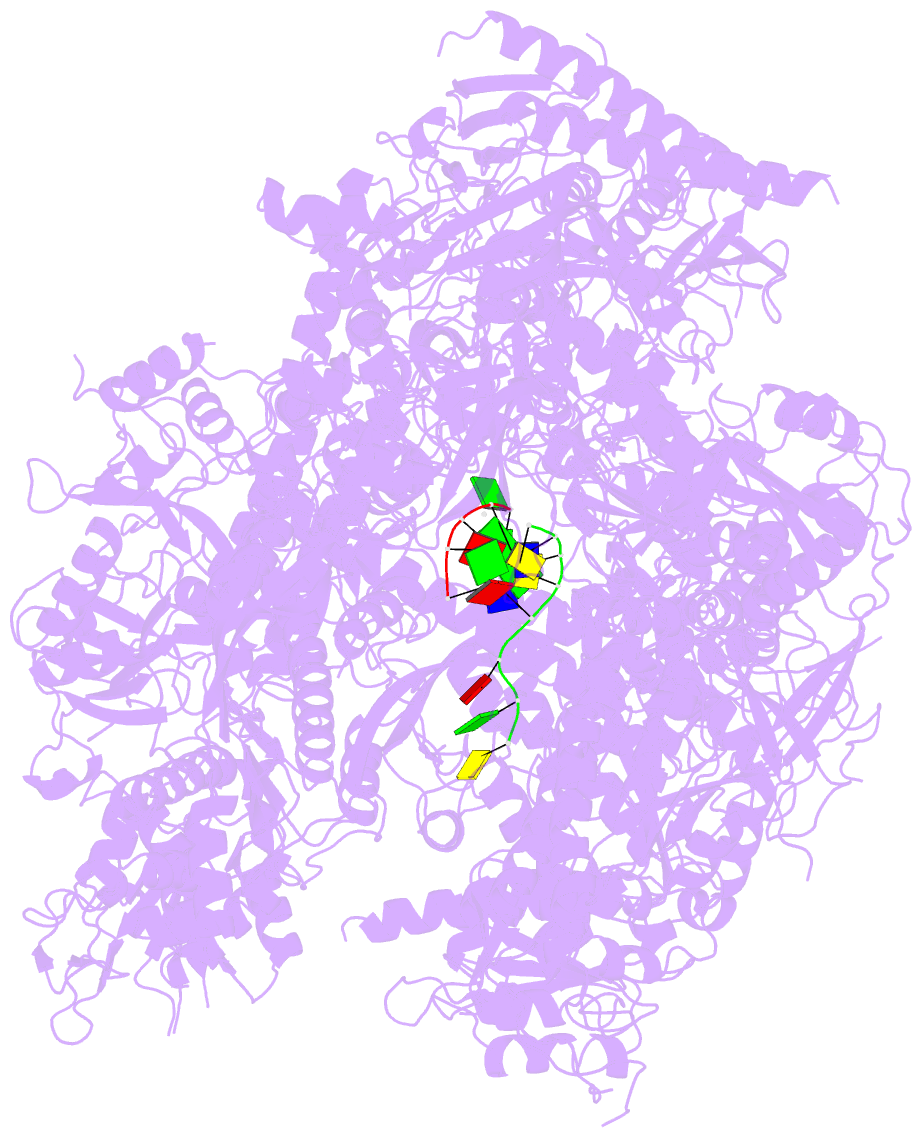
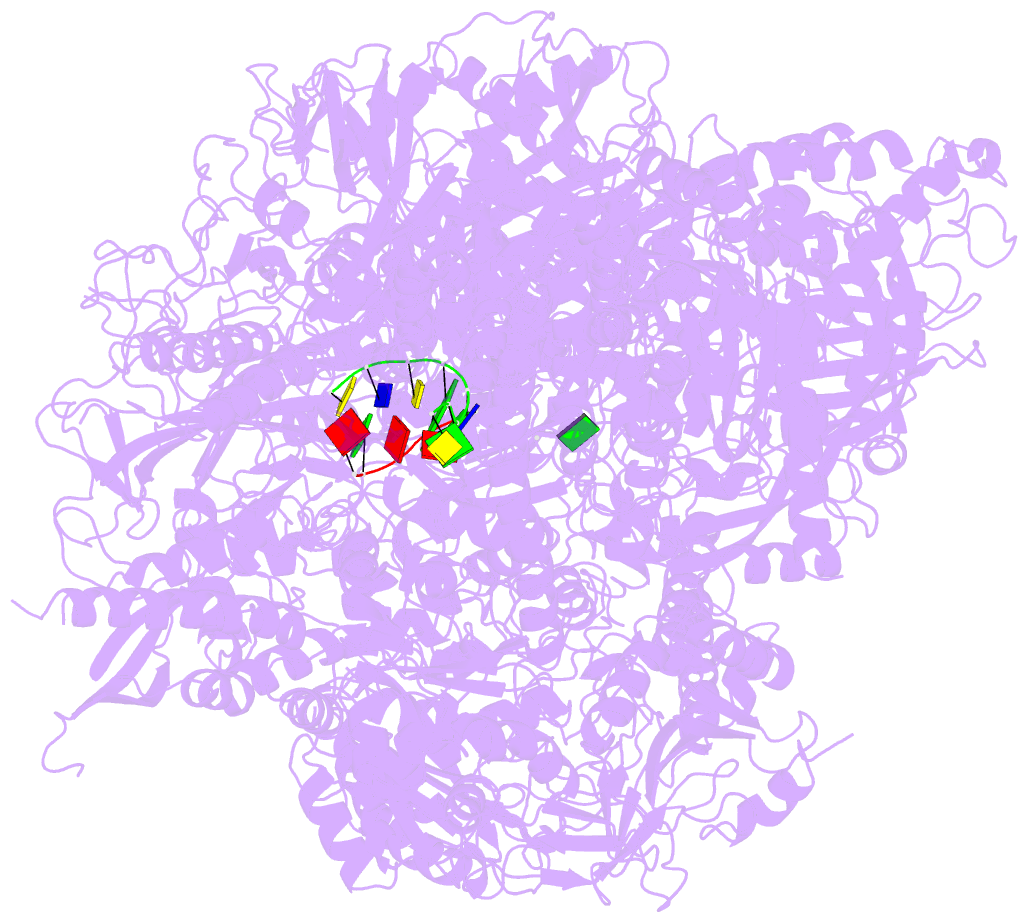
- Summary
- RNA polymerase ii initiation complex with a 5-nt 3'-deoxy RNA soaked with gtp
- Reference
- Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD (2011): "Initiation complex structure and promoter proofreading." Science, 333, 633-637. doi: 10.1126/science.1206629.
- Abstract
- The initiation of transcription by RNA polymerase II is a multistage process. X-ray crystal structures of transcription complexes containing short RNAs reveal three structural states: one with 2- and 3-nucleotide RNAs, in which only the 3'-end of the RNA is detectable; a second state with 4- and 5-nucleotide RNAs, with an RNA-DNA hybrid in a grossly distorted conformation; and a third state with RNAs of 6 nucleotides and longer, essentially the same as a stable elongating complex. The transition from the first to the second state correlates with a markedly reduced frequency of abortive initiation. The transition from the second to the third state correlates with partial "bubble collapse" and promoter escape. Polymerase structure is permissive for abortive initiation, thereby setting a lower limit on polymerase-promoter complex lifetime and allowing the dissociation of nonspecific complexes. Abortive initiation may be viewed as promoter proofreading, and the structural transitions as checkpoints for promoter control.