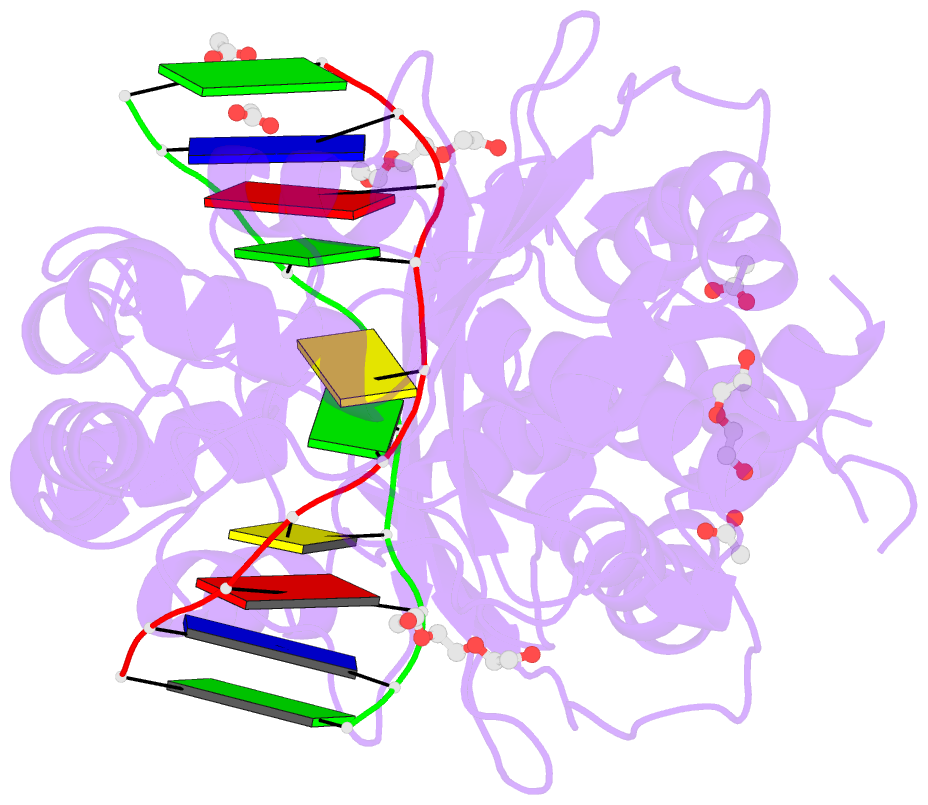
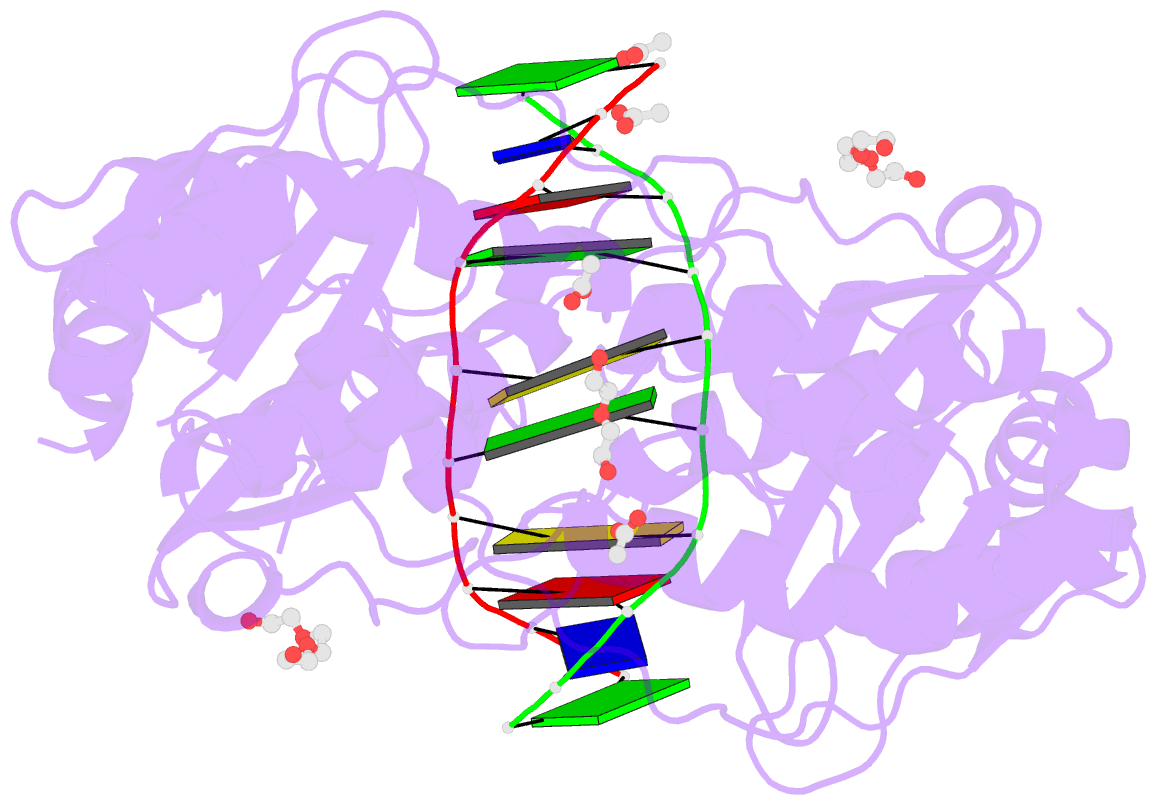
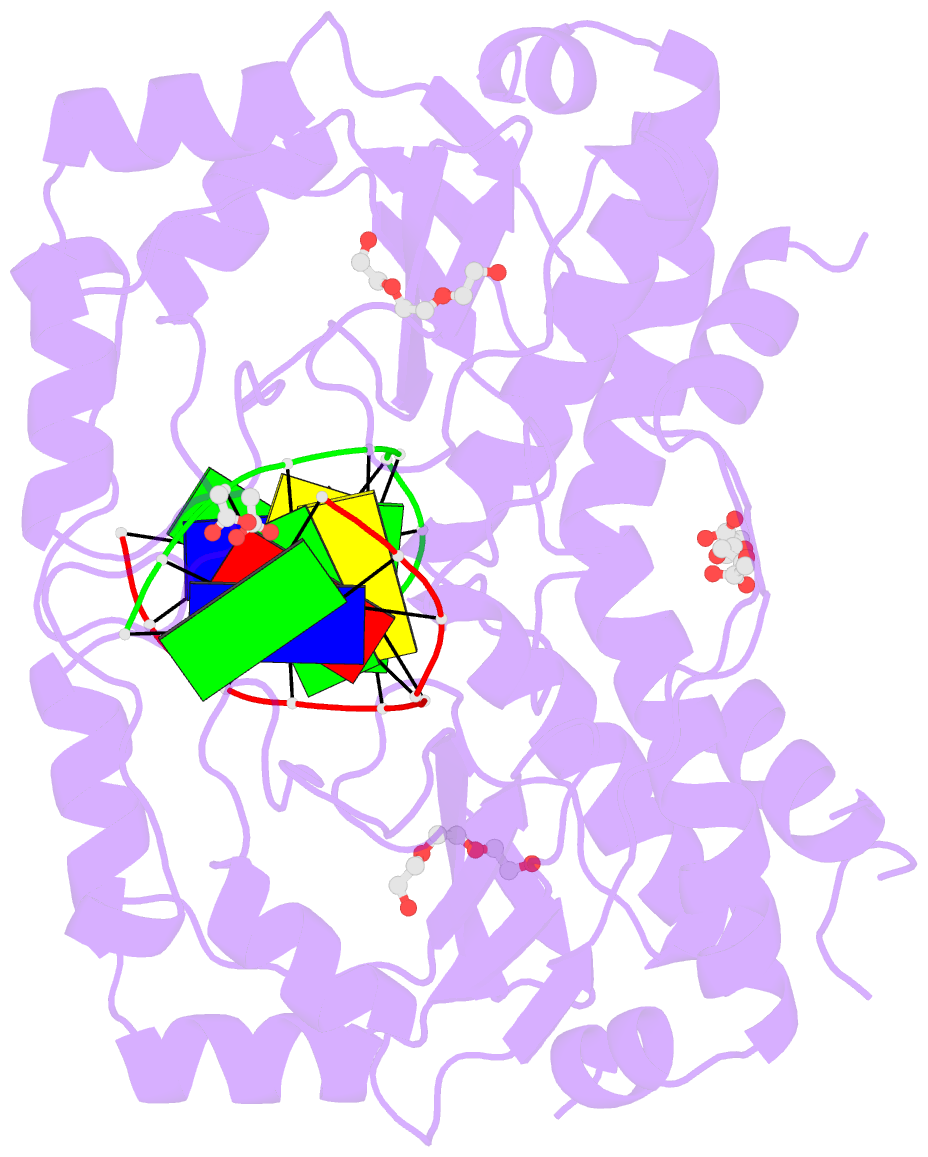
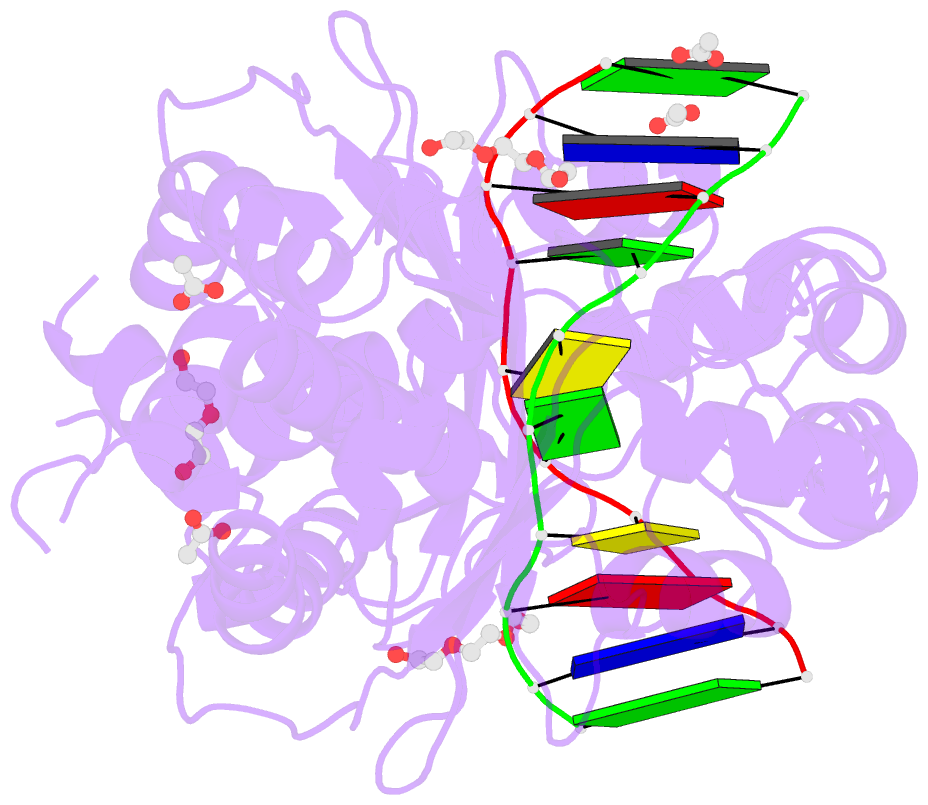
Summary information and primary citation
- PDB-id
- 3ndh; DSSR-derived features in text and JSON formats
- Class
- hydrolase-DNA
- Method
- X-ray (1.3 Å)
- Summary
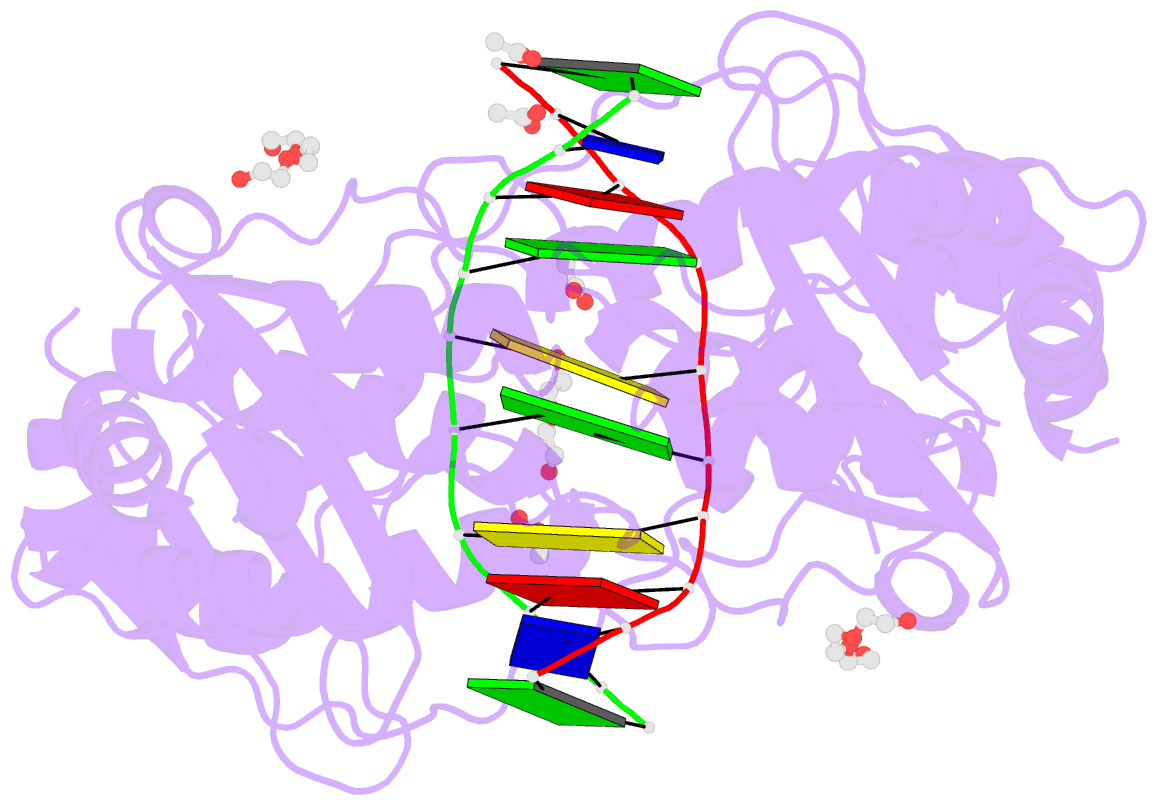
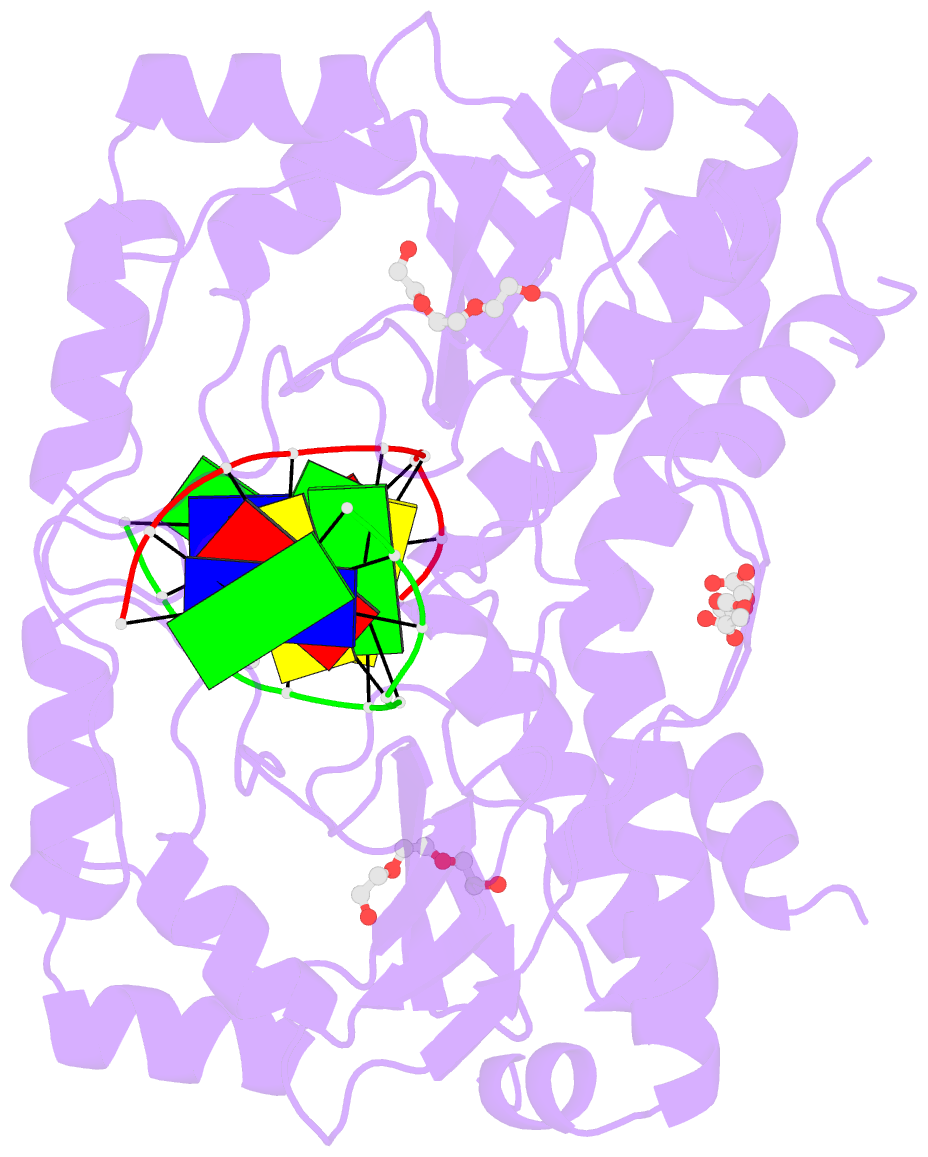
- Restriction endonuclease in complex with substrate DNA
- Reference
- Firczuk M, Wojciechowski M, Czapinska H, Bochtler M (2011): "DNA intercalation without flipping in the specific ThaI-DNA complex." Nucleic Acids Res., 39, 744-754. doi: 10.1093/nar/gkq834.
- Abstract
- The PD-(D/E)XK type II restriction endonuclease ThaI cuts the target sequence CG/CG with blunt ends. Here, we report the 1.3 Å resolution structure of the enzyme in complex with substrate DNA and a sodium or calcium ion taking the place of a catalytic magnesium ion. The structure identifies Glu54, Asp82 and Lys93 as the active site residues. This agrees with earlier bioinformatic predictions and implies that the PD and (D/E)XK motifs in the sequence are incidental. DNA recognition is very unusual: the two Met47 residues of the ThaI dimer intercalate symmetrically into the CG steps of the target sequence. They approach the DNA from the minor groove side and penetrate the base stack entirely. The DNA accommodates the intercalating residues without nucleotide flipping by a doubling of the CG step rise to twice its usual value, which is accompanied by drastic unwinding. Displacement of the Met47 side chains from the base pair midlines toward the downstream CG steps leads to large and compensating tilts of the first and second CG steps. DNA intercalation by ThaI is unlike intercalation by HincII, HinP1I or proteins that bend or repair DNA.