Summary information and primary citation
- PDB-id
- 3iv5; DSSR-derived features in text and JSON formats
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.9 Å)
- Summary
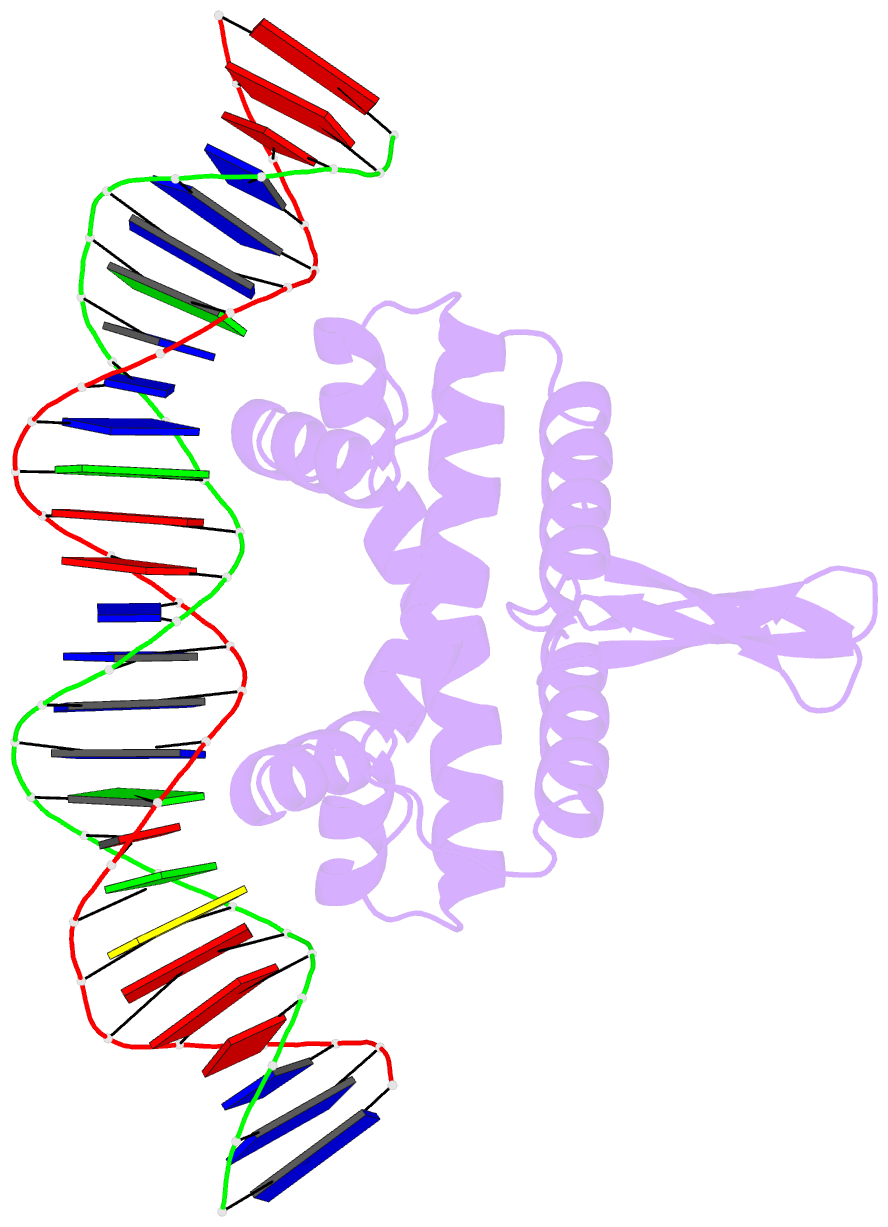
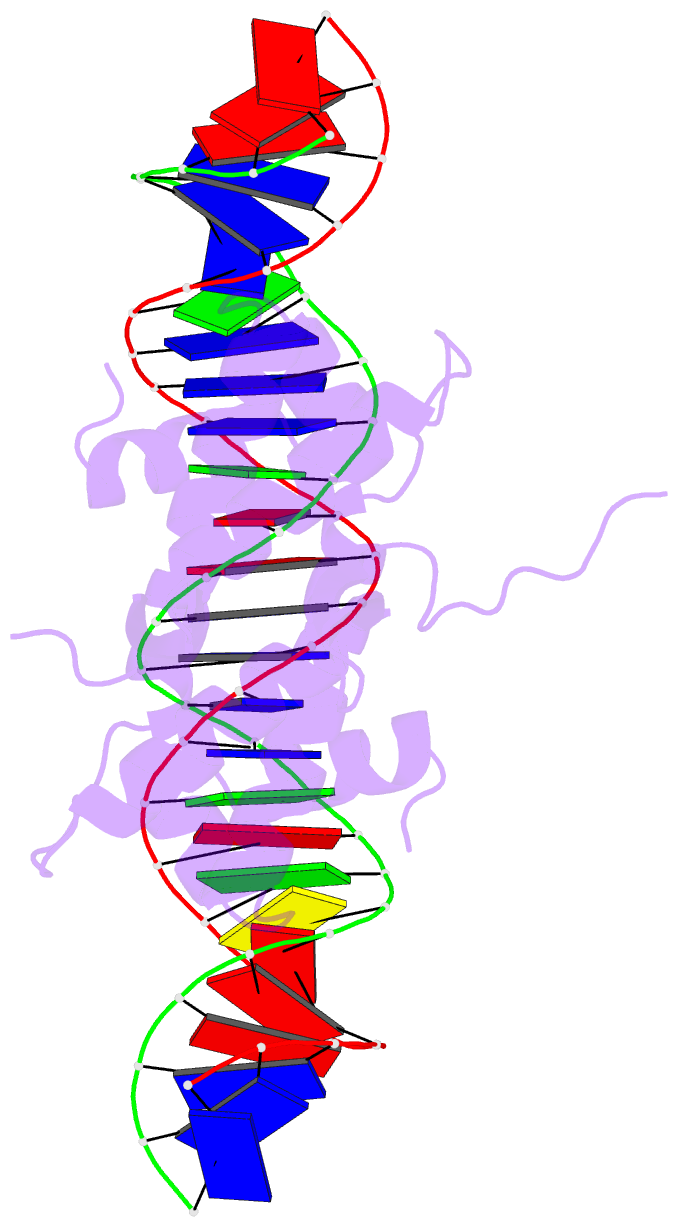
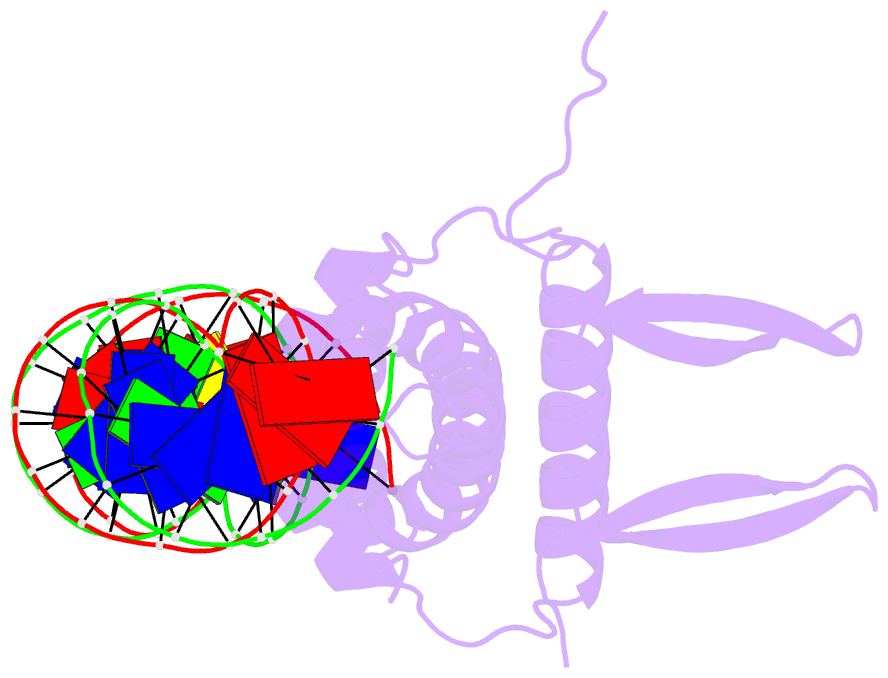
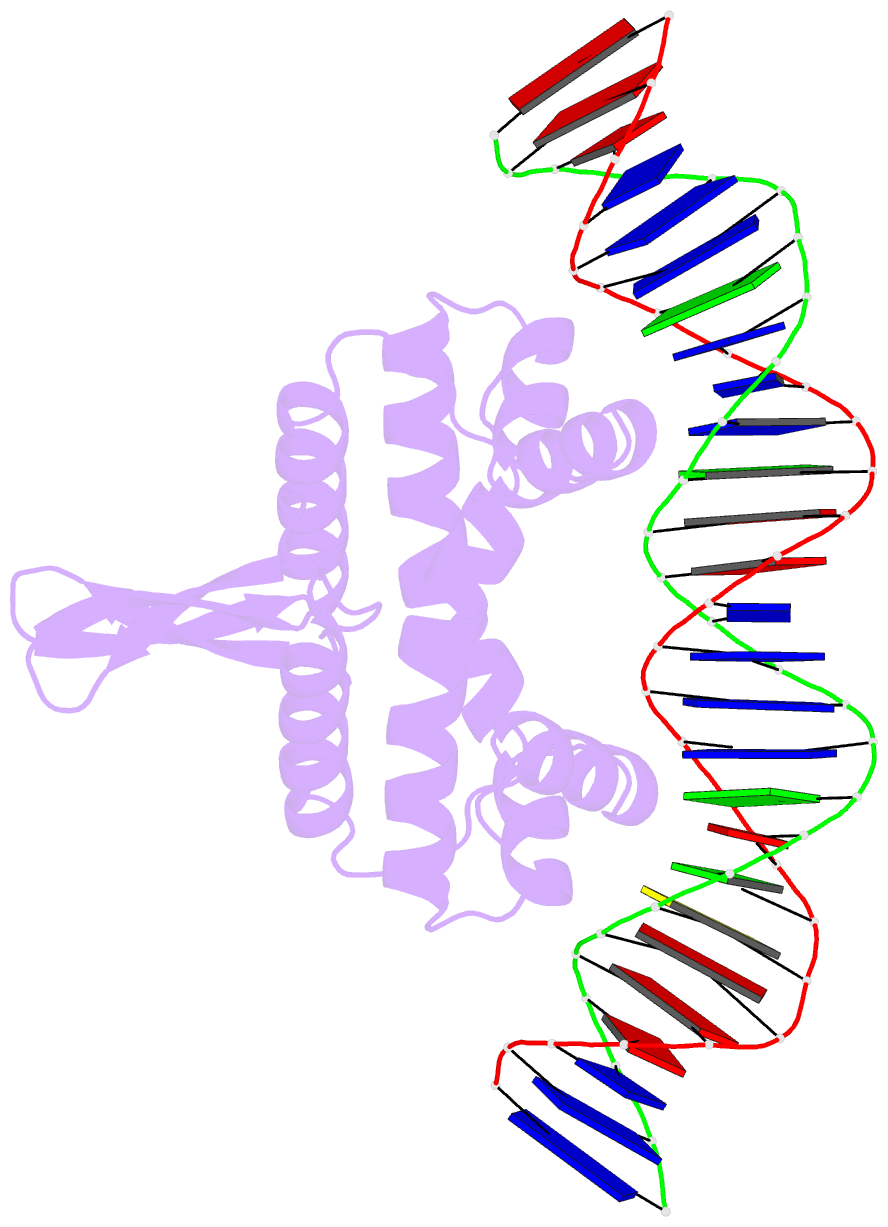
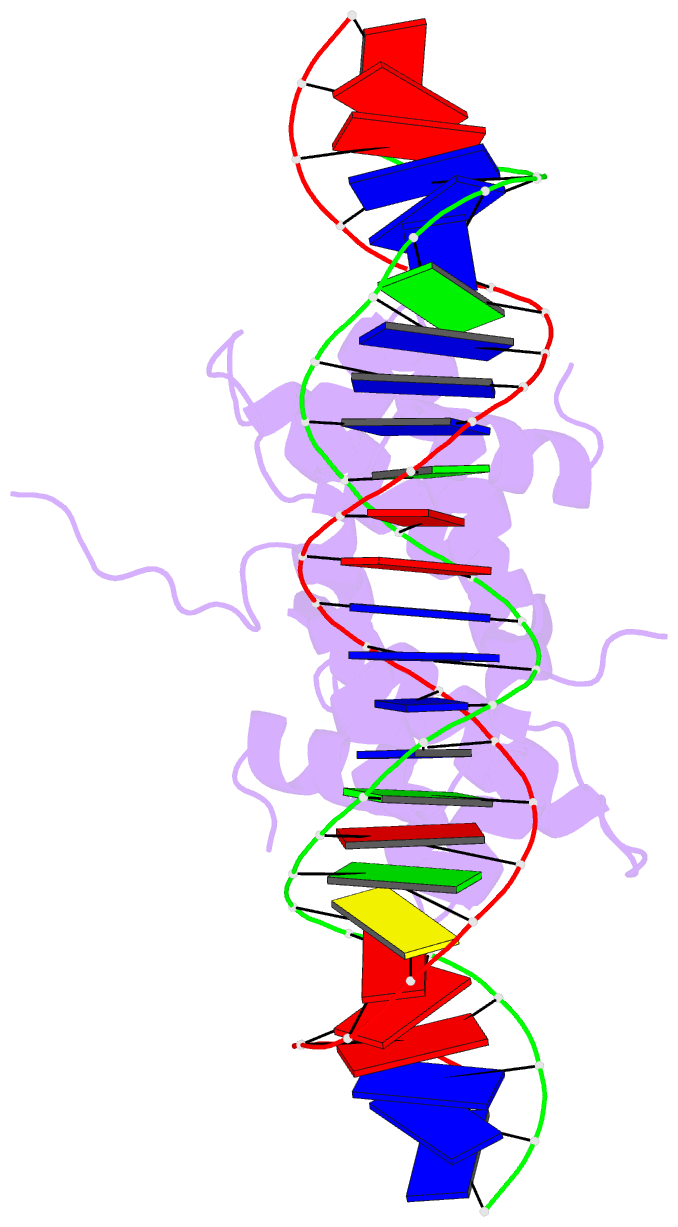
- Crystal structure of fis bound to 27 bp optimal binding sequence f1
- Reference
- Stella S, Cascio D, Johnson RC (2010): "The shape of the DNA minor groove directs binding by the DNA-bending protein Fis." Genes Dev., 24, 814-826. doi: 10.1101/gad.1900610.
- Abstract
- The bacterial nucleoid-associated protein Fis regulates diverse reactions by bending DNA and through DNA-dependent interactions with other control proteins and enzymes. In addition to dynamic nonspecific binding to DNA, Fis forms stable complexes with DNA segments that share little sequence conservation. Here we report the first crystal structures of Fis bound to high- and low-affinity 27-base-pair DNA sites. These 11 structures reveal that Fis selects targets primarily through indirect recognition mechanisms involving the shape of the minor groove and sequence-dependent induced fits over adjacent major groove interfaces. The DNA shows an overall curvature of approximately 65 degrees , and the unprecedented close spacing between helix-turn-helix motifs present in the apodimer is accommodated by severe compression of the central minor groove. In silico DNA structure models show that only the roll, twist, and slide parameters are sufficient to reproduce the changes in minor groove widths and recreate the curved Fis-bound DNA structure. Models based on naked DNA structures suggest that Fis initially selects DNA targets with intrinsically narrow minor grooves using the separation between helix-turn-helix motifs in the Fis dimer as a ruler. Then Fis further compresses the minor groove and bends the DNA to generate the bound structure.