Summary information and primary citation
- PDB-id
- 3au6; DSSR-derived features in text and JSON formats
- Class
- transferase-DNA
- Method
- X-ray (3.3 Å)
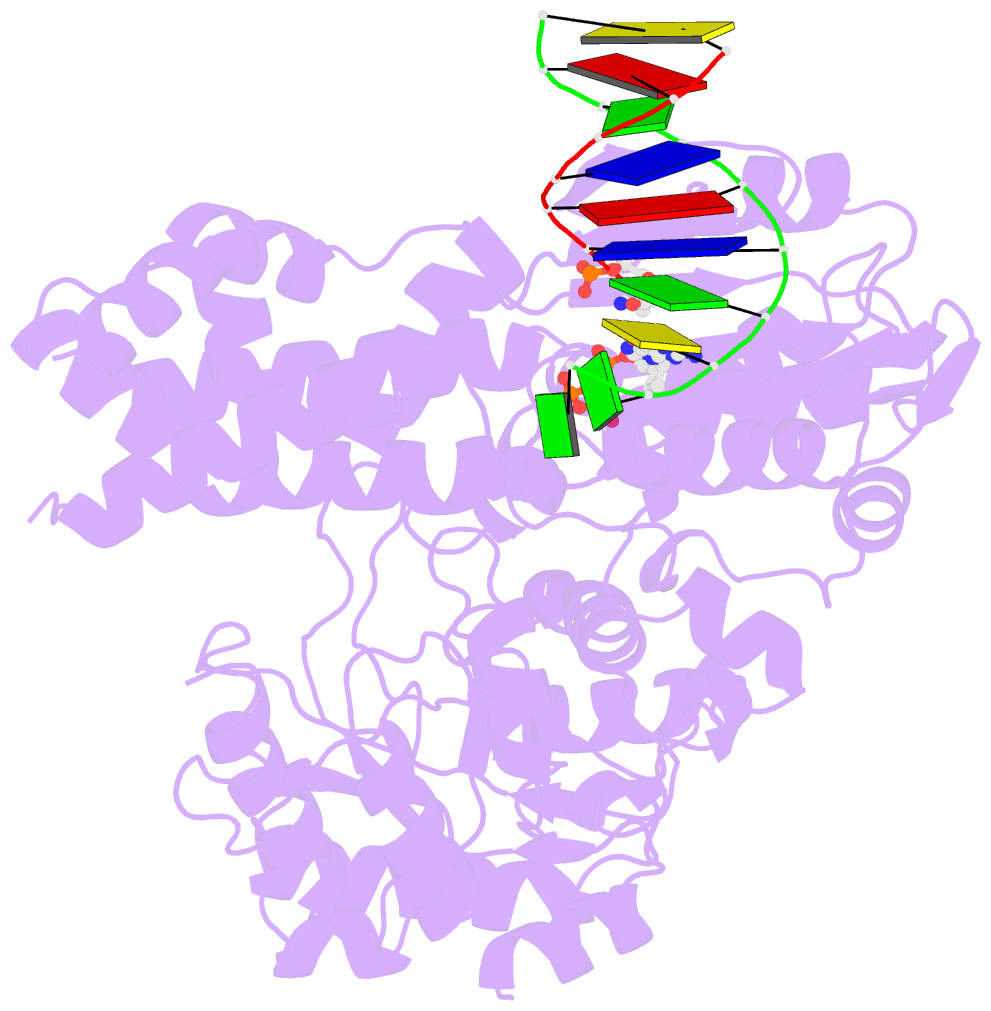
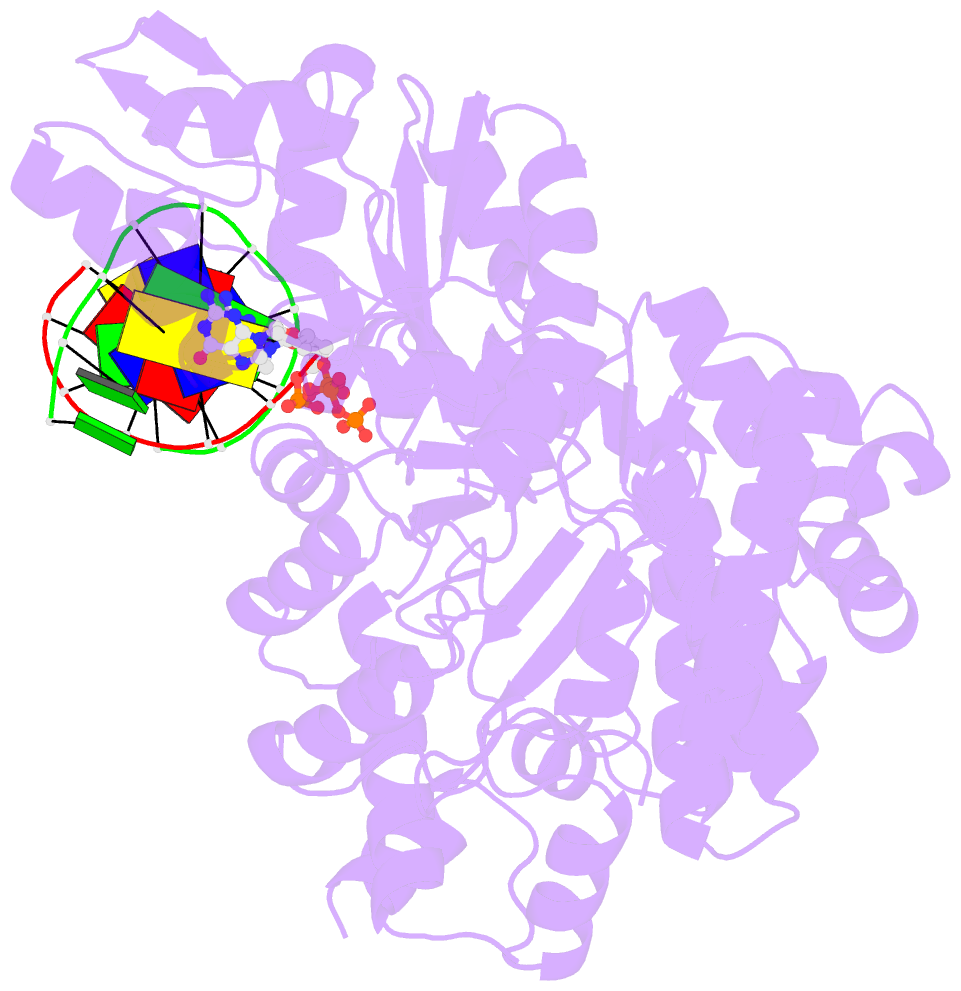
- Summary
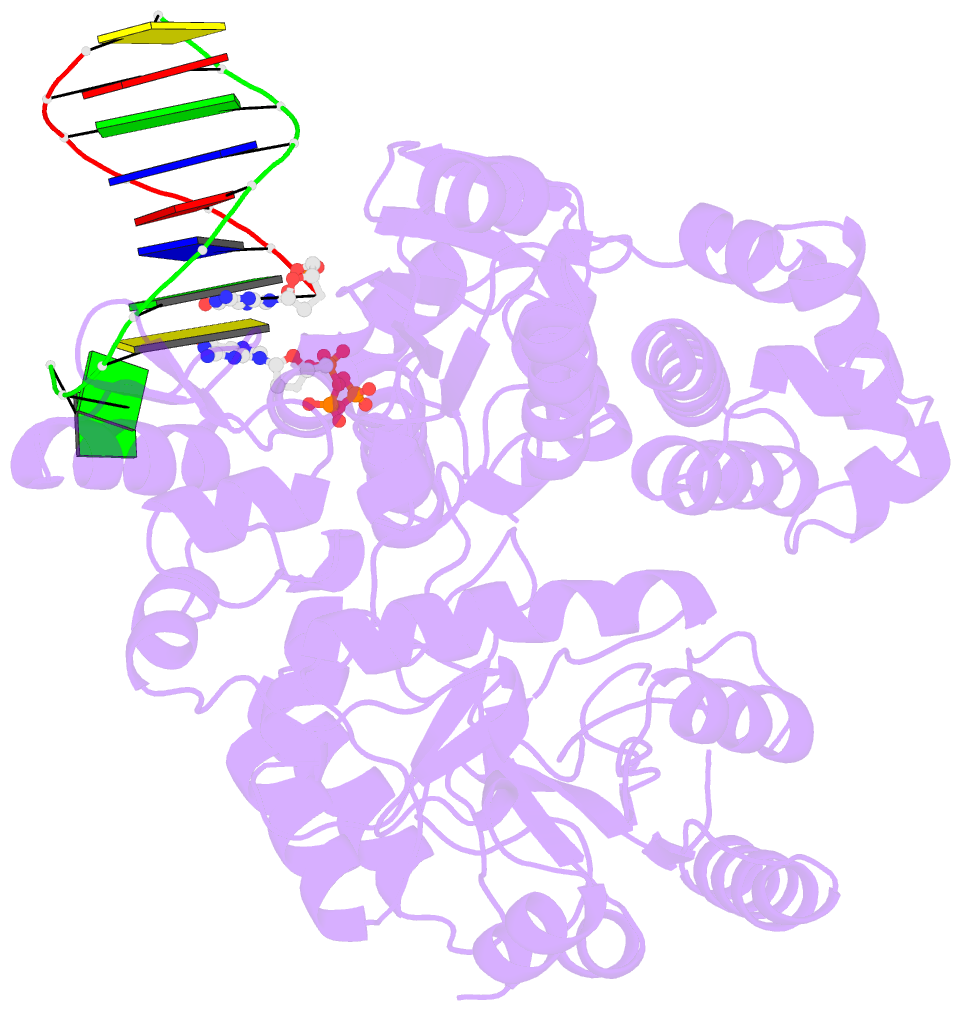
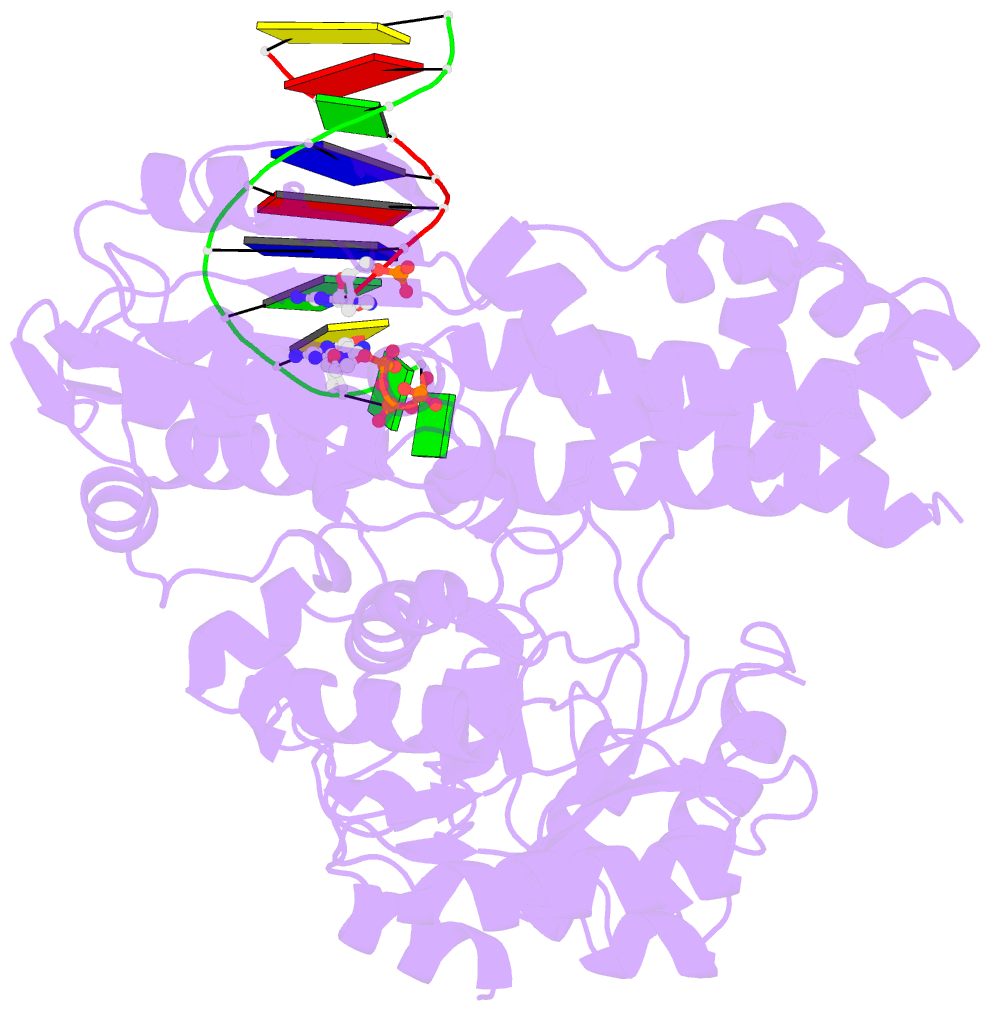
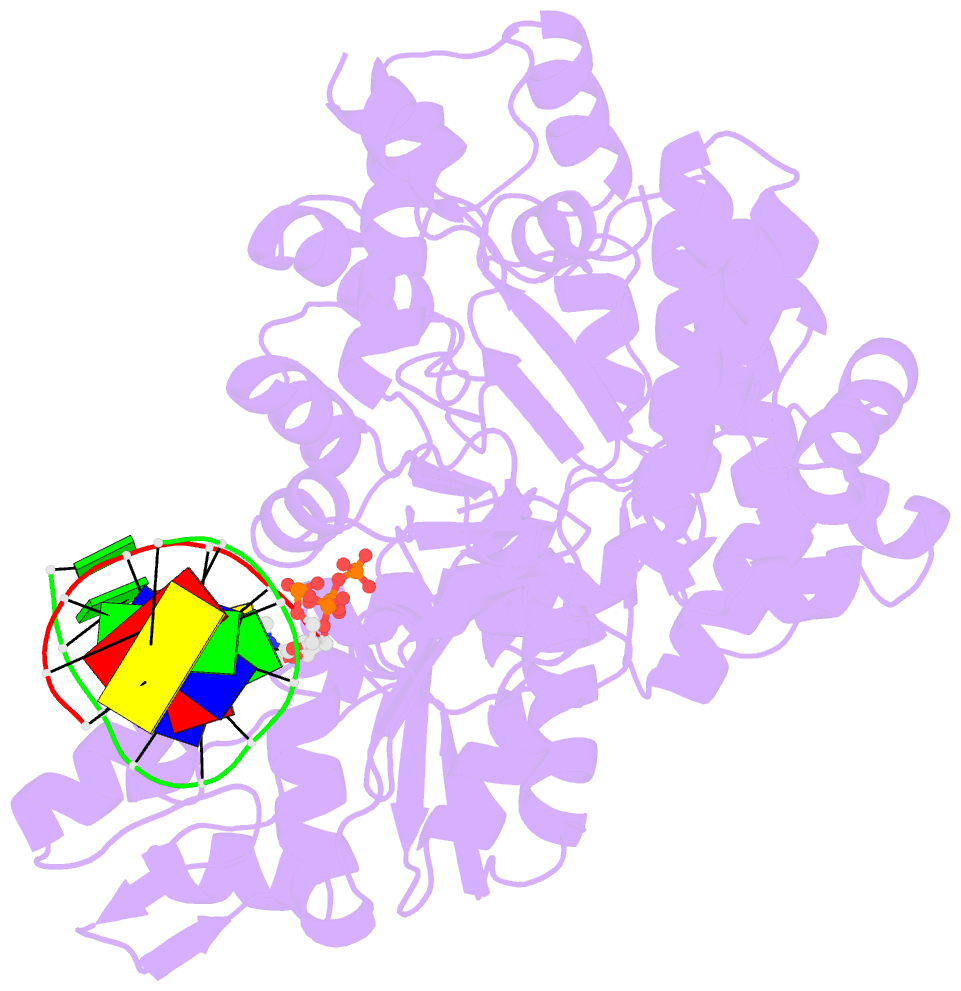
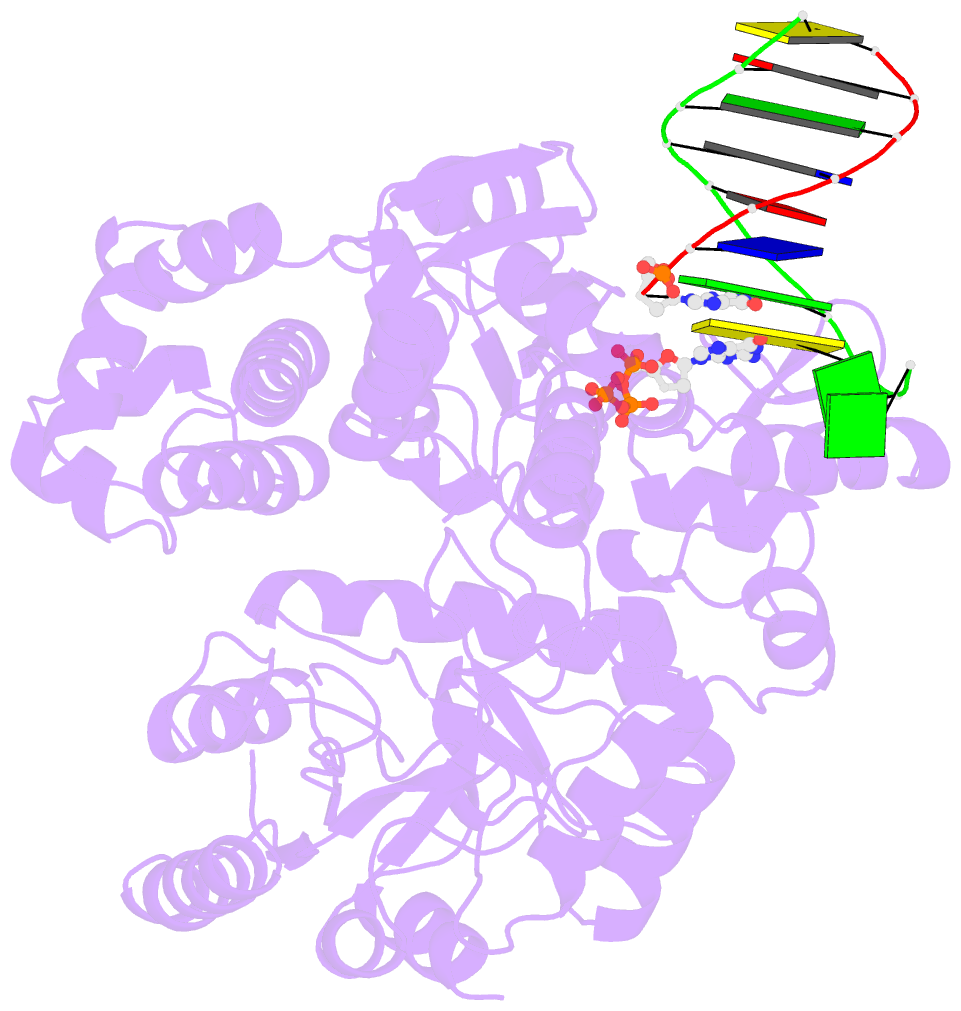
- DNA polymerase x from thermus thermophilus hb8 ternary complex with primer-template DNA and ddgtp
- Reference
- Nakane S, Ishikawa H, Nakagawa N, Kuramitsu S, Masui R (2012): "The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA." J.Mol.Biol., 417, 179-196. doi: 10.1016/j.jmb.2012.01.025.
- Abstract
- DNA with single-nucleotide (1-nt) gaps can arise during various DNA processing events. These lesions are repaired by X-family DNA polymerases (PolXs) with high gap-filling activity. Some PolXs can bind productively to dNTPs in the absence of DNA and fill these 1-nt gaps. Although PolXs have a crucial role in efficient gap filling, currently, little is known of the kinetic and structural details of their productive dNTP binding. Here, we show that Thermus thermophilus HB8 PolX (ttPolX) had strong binding affinity for Mg(2+)-dNTPs in the absence of DNA and that it follows a Theorell-Chance (hit-and-run) mechanism with nucleotide binding first. Comparison of the intermediate crystal structures of ttPolX in a binary complex with dGTP and in a ternary complex with 1-nt gapped DNA and Mg(2+)-ddGTP revealed that the conformation of the incoming nucleotide depended on whether or not DNA was present. Furthermore, the Lys263 residue located between two guanosine conformations was essential to the strong binding affinity of the enzyme. The ability to bind to either syn-dNTP or anti-dNTP and the involvement of a Theorell-Chance mechanism are key aspects of the strong nucleotide-binding and efficient gap-filling activities of ttPolX.