Summary information and primary citation
- PDB-id
- 2vum; DSSR-derived features in text and JSON formats
- Class
- transferase
- Method
- X-ray (3.4 Å)
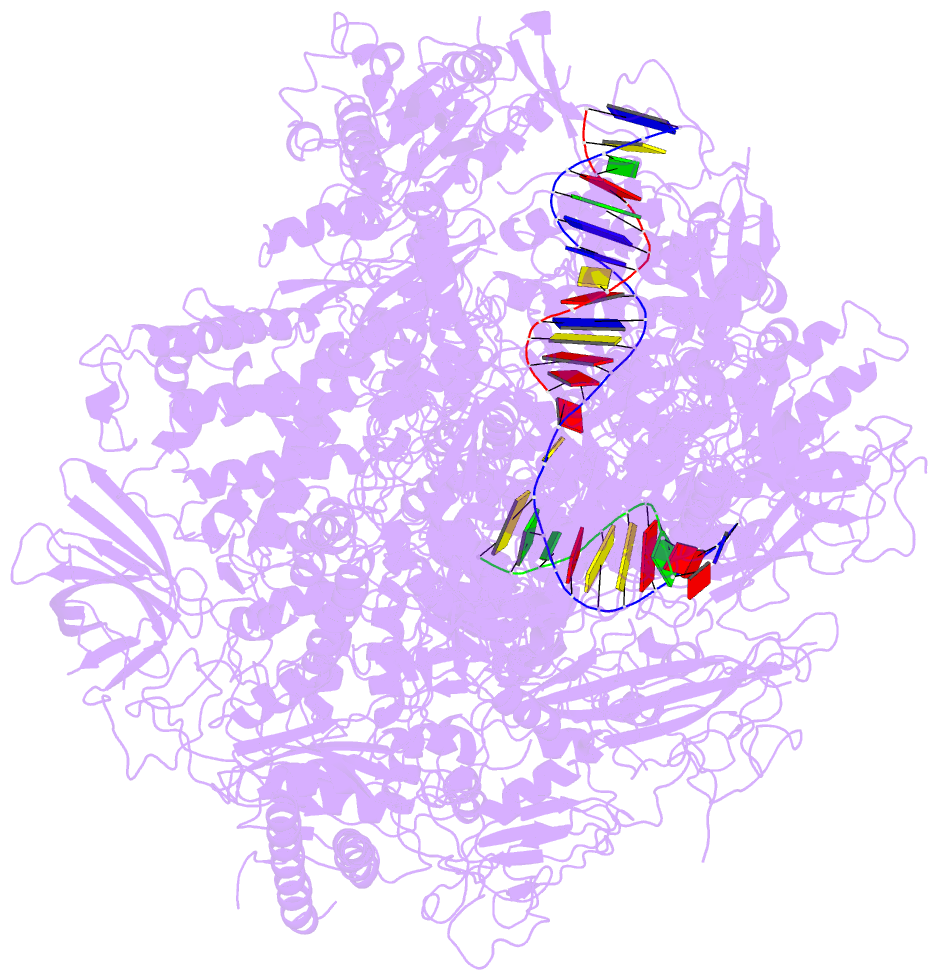
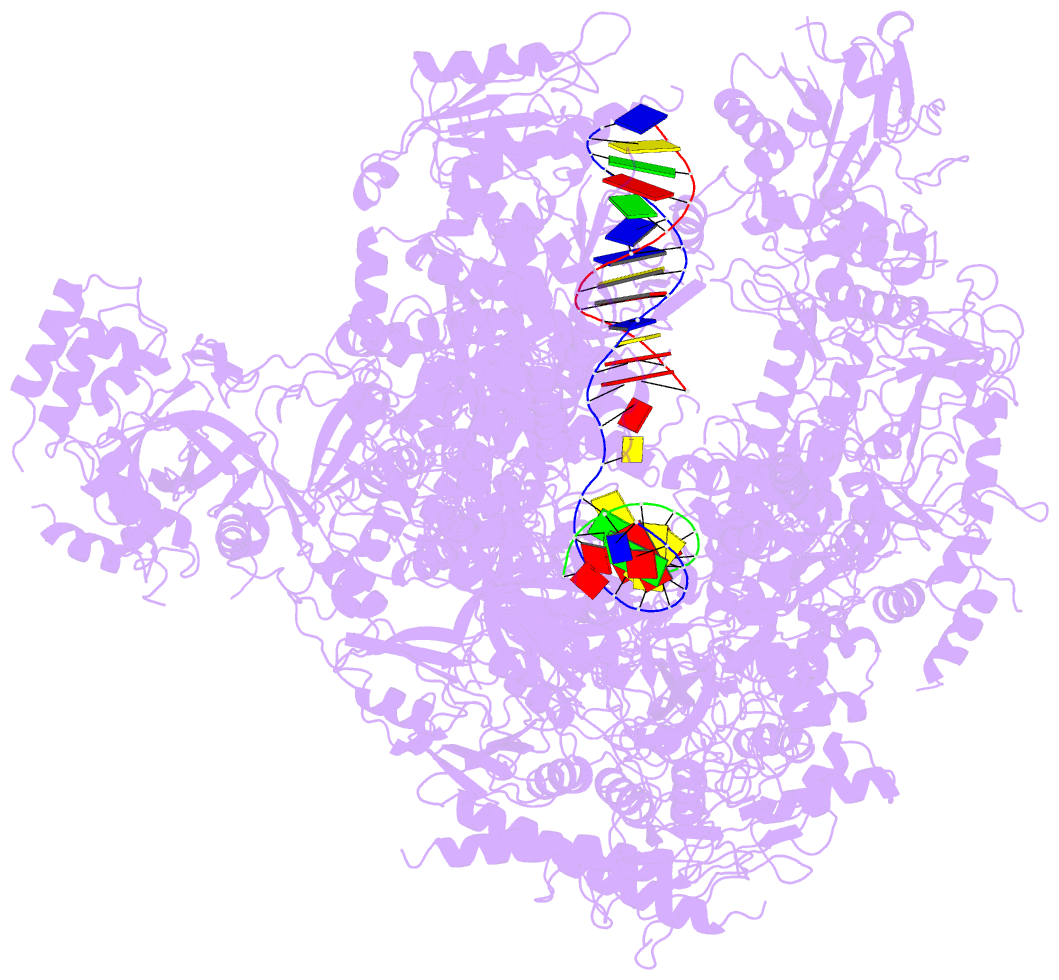
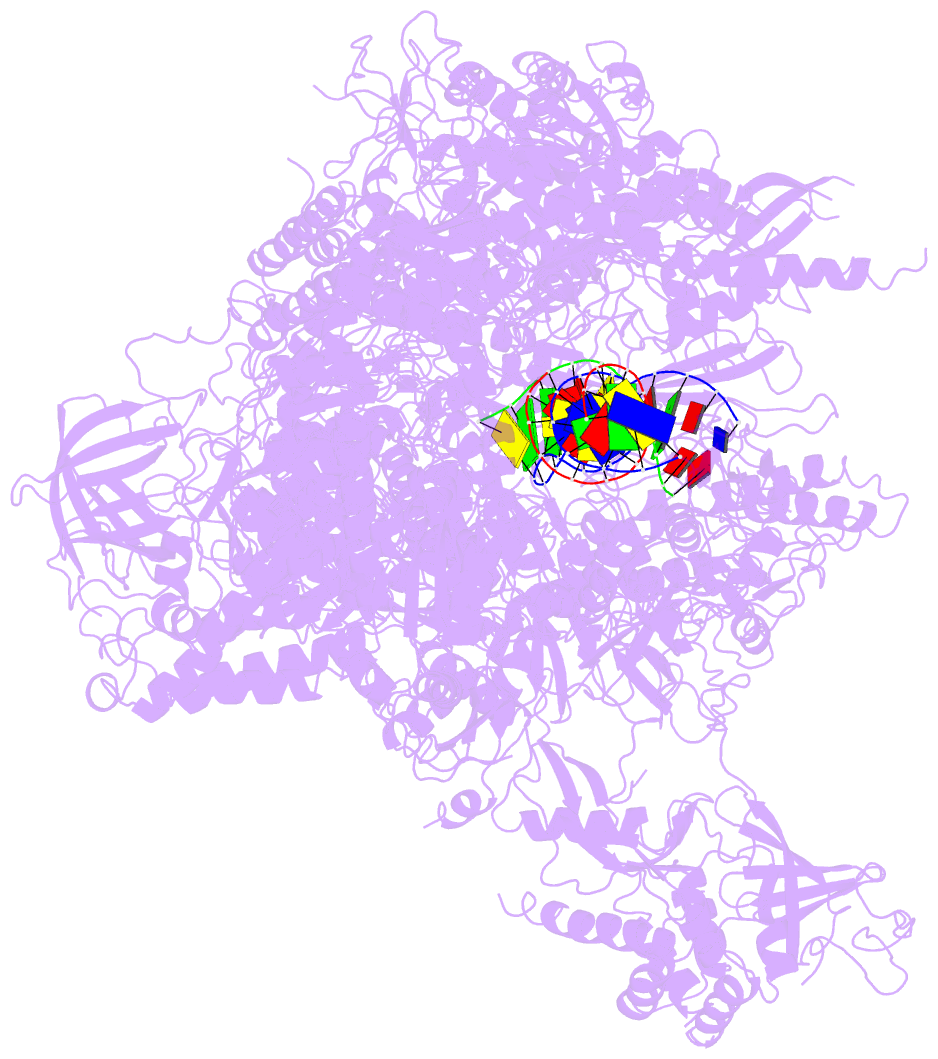
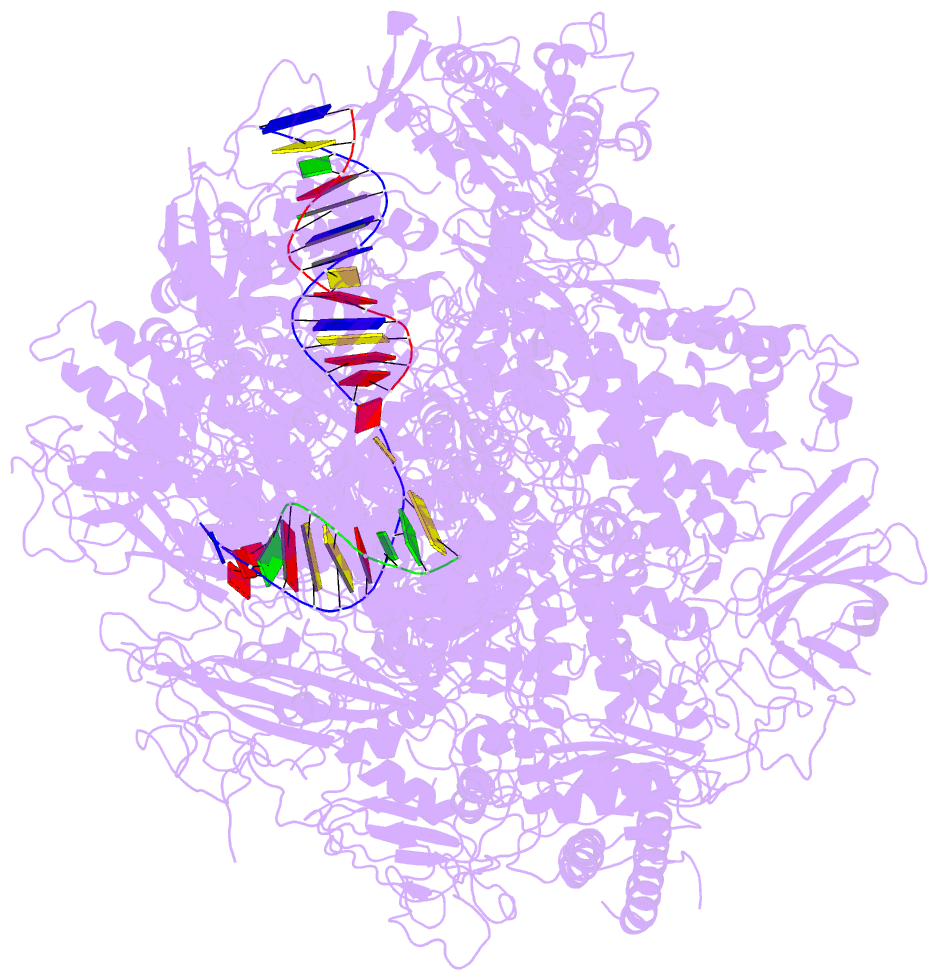
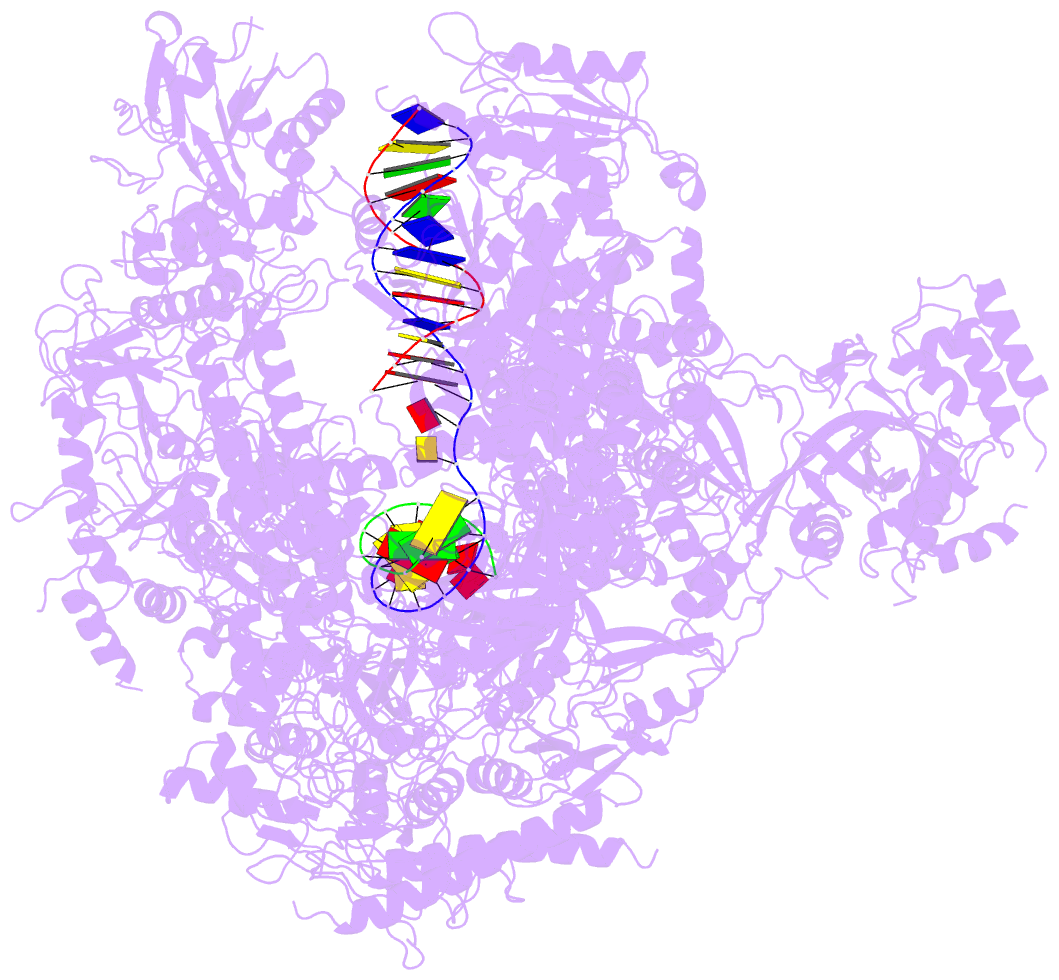
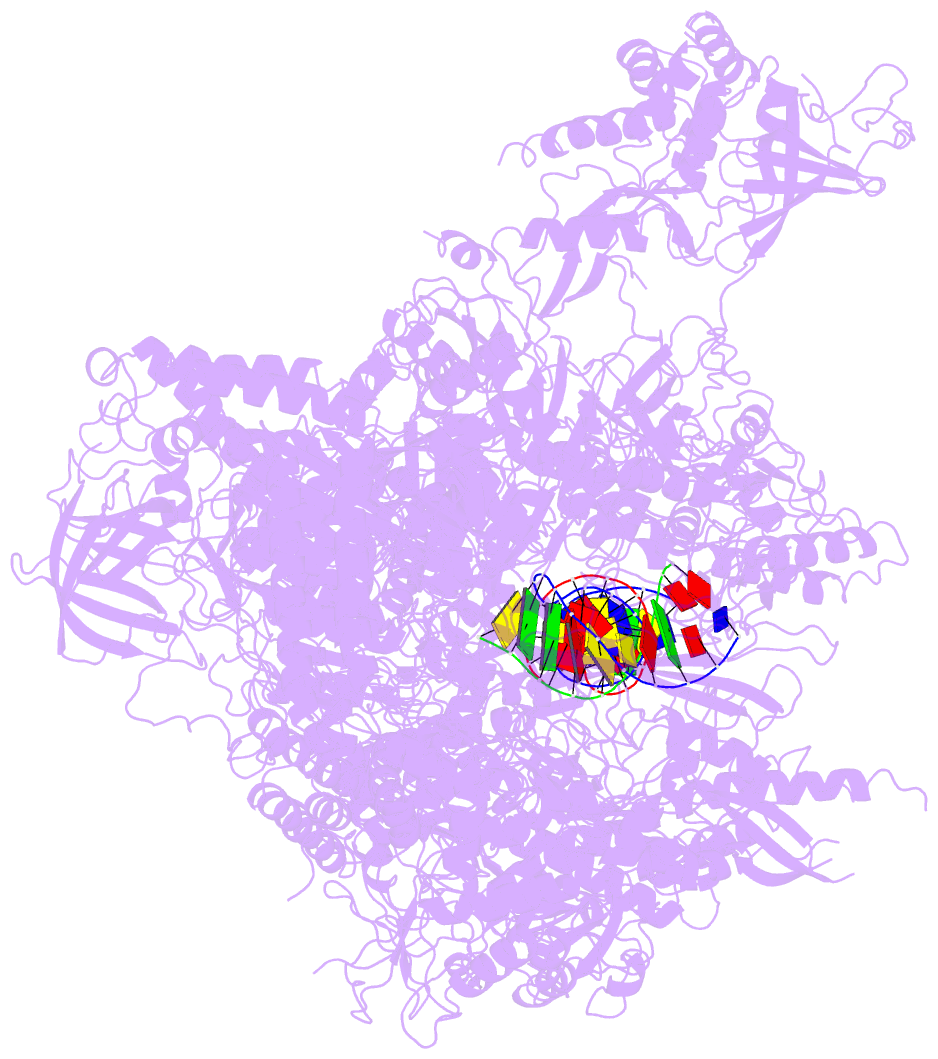
- Summary
- Alpha-amanitin inhibited complete RNA polymerase ii elongation complex
- Reference
- Brueckner F, Cramer P (2008): "Structural Basis of Transcription Inhibition by Alpha-Amanitin and Implications for RNA Polymerase II Translocation." Nat.Struct.Mol.Biol., 15, 811. doi: 10.1038/NSMB.1458.
- Abstract
- To study how RNA polymerase II translocates after nucleotide incorporation, we prepared elongation complex crystals in which pre- and post-translocation states interconvert. Crystal soaking with the inhibitor alpha-amanitin locked the elongation complex in a new state, which was refined at 3.4-A resolution and identified as a possible translocation intermediate. The DNA base entering the active site occupies a 'pretemplating' position above the central bridge helix, which is shifted and occludes the templating position. A leucine residue in the trigger loop forms a wedge at the shifted bridge helix, but moves by 13 A to close the active site during nucleotide incorporation. Our results support a Brownian ratchet mechanism that involves swinging of the trigger loop between open, wedged and closed positions, and suggest that alpha-amanitin impairs nucleotide incorporation and translocation by trapping the trigger loop and bridge helix.