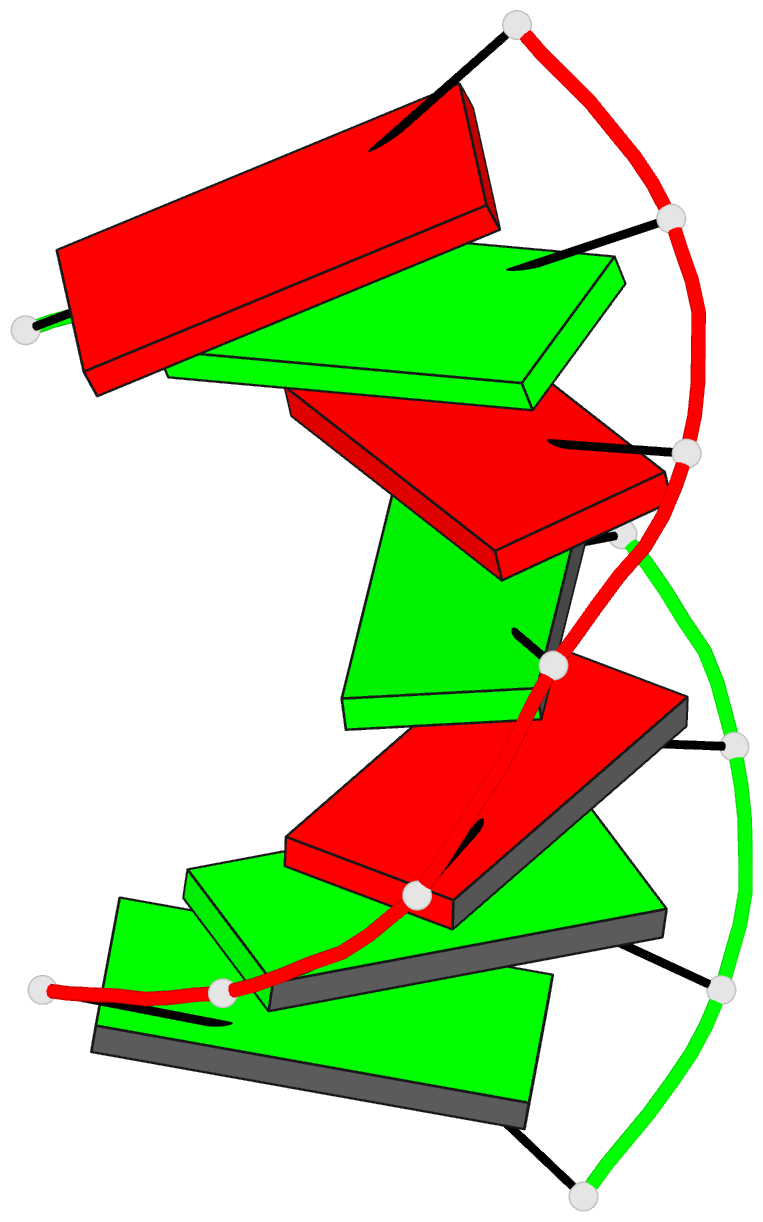
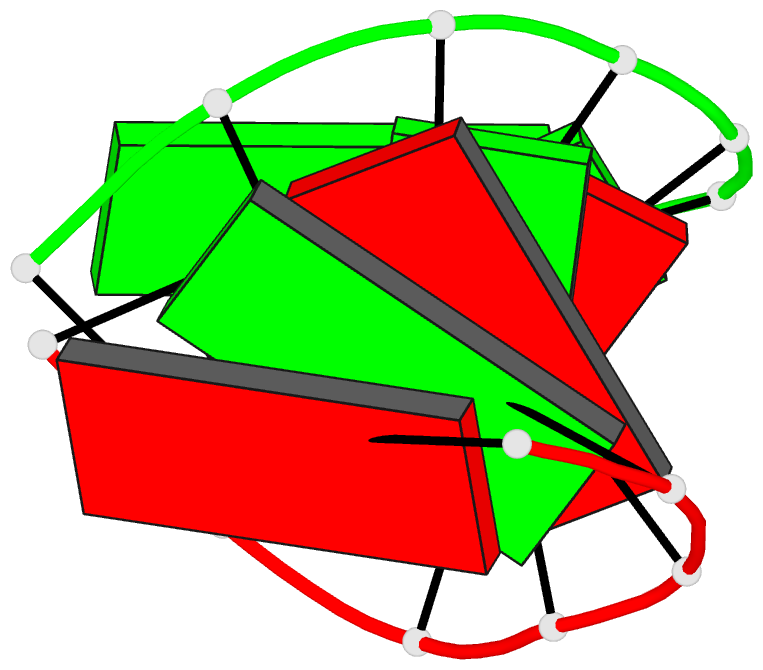
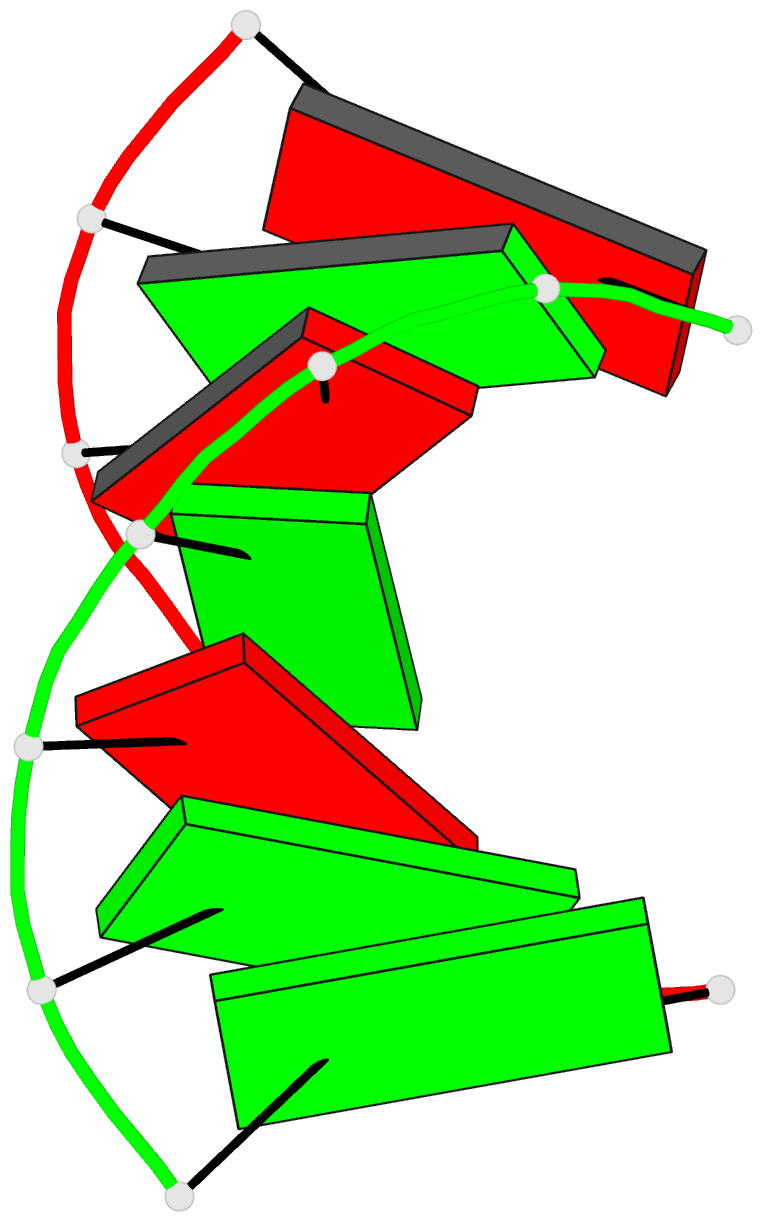
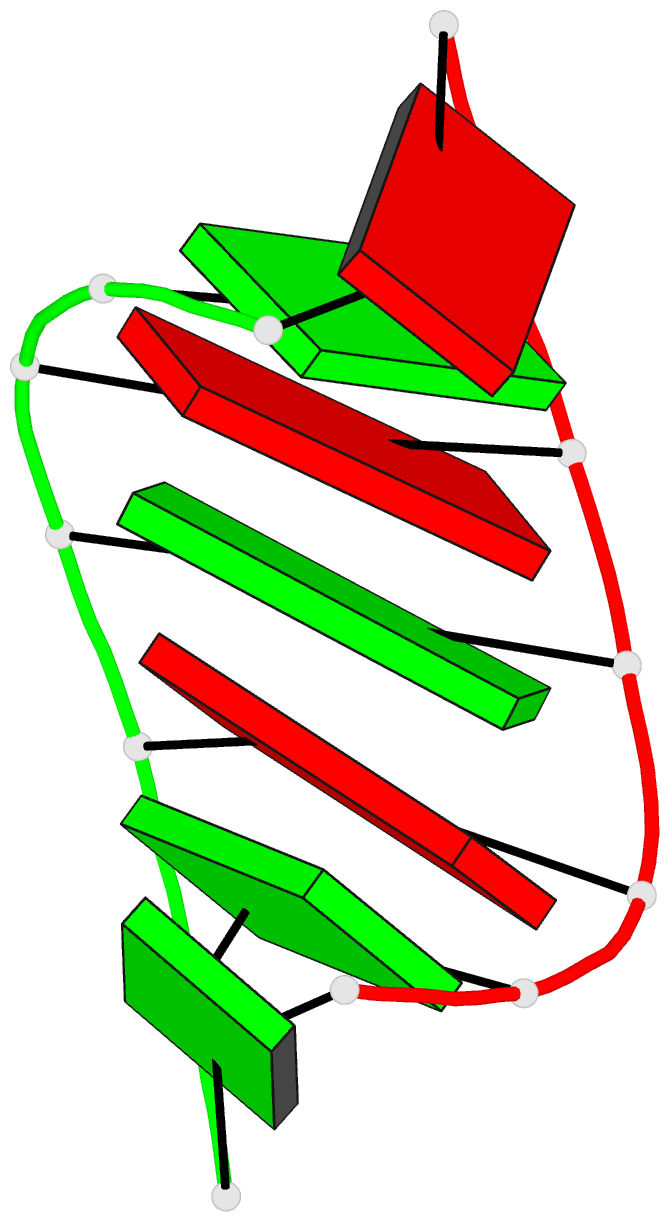
Summary information and primary citation
- PDB-id
-
2v6w;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (1.8 Å)
- Summary
- Trnaser acceptor stem: conformation and hydration of a
microhelix in a crystal structure at 1.8 angstrom
resolution
- Reference
-
Forster C, Brauer ABE, Brode S, Furste JP, Betzel C,
Erdmann VA (2007): "Trnaser
Acceptor Stem: Conformation and Hydration of a Microhelix
in a Crystal Structure at 1.8 A Resolution." Acta
Crystallogr.,Sect.D, 63, 1154. doi:
10.1107/S0907444907045271.
- Abstract
- The crystal structure of a serine-specific tRNA
acceptor-stem microhelix, the binding site for the
seryl-tRNA synthetase, was solved by X-ray analysis. This
seven-base-pair tRNA(Ser) microhelix forms endless rows of
helices in the crystal lattice, with two helices stacking
'head-to-head' onto each other, resulting in an
intermolecular guanosine stacking of the first purine
nucleotides at the 5'-strands of the tRNA(Ser)
microhelices. A network of 75 water loci could be
associated with each RNA duplex. Unusual local geometric
backbone parameters could be detected in the region of the
G4 phosphate located in the 5'-strand of the helix, which
lead to a ;kink' in this region and to an irregularly bent
helix. The role of the specific hydration pattern and of
the irregular conformation of the tRNA(Ser) acceptor-stem
helix is discussed and summarized.